A CD147-targeted small-molecule inhibitor potentiates gemcitabine efficacy by triggering ferroptosis in pancreatic ductal adenocarcinoma
- PMID: 40834852
- PMCID: PMC12432379
- DOI: 10.1016/j.xcrm.2025.102292
A CD147-targeted small-molecule inhibitor potentiates gemcitabine efficacy by triggering ferroptosis in pancreatic ductal adenocarcinoma
Abstract
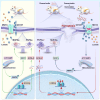
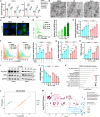
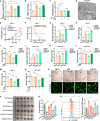
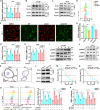
CD147 has emerged as a promising tumor-specific therapeutic target. Identifying small-molecule inhibitors that promote its proteolysis represents a critical step toward advancing clinical translation, while elucidating its mechanisms of action could further accelerate this process. In this study, we identify dracorhodin perchlorate (DP) as a potent CD147 inhibitor that induces autophagy-dependent degradation. DP significantly inhibits cell proliferation and enhances sensitivity to gemcitabine in pancreatic cancer cells. Mechanistically, CD147 inhibition upregulates acyl-CoA synthetase long-chain family member 4 (ACSL4) expression through H3K9 lactylation and suppresses the sterol regulatory element-binding protein 1 (SREBP1)/stearoyl-CoA desaturase-1 (SCD1) signaling pathway, collectively disrupting the balance of polyunsaturated and monounsaturated fatty acids, ultimately triggering ferroptosis. The combination of DP and gemcitabine demonstrates remarkable synergistic anti-tumor effects in orthotopic xenograft models, spontaneous KPC mouse models, and patient-derived organoid (PDO) and xenograft (PDX) models. In conclusion, this study reveals a mechanism by which CD147 regulates ferroptosis and supports combining DP with gemcitabine as a therapeutic strategy to improve patient outcomes in pancreatic ductal adenocarcinoma.
Keywords: ACSL4; CD147; H3K9la; dracorhodin; ferroptosis; gemcitabine; pancreatic ductal adenocarcinoma.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Huang Q., Li J., Xing J., Li W., Li H., Ke X., Zhang J., Ren T., Shang Y., Yang H., et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J. Hepatol. 2014;61:859–866. doi: 10.1016/j.jhep.2014.04.035. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

