Intracristal space proteome mapping using super-resolution proximity labeling with isotope-coded probes
- PMID: 40835710
- PMCID: PMC12368266
- DOI: 10.1038/s41467-025-62756-0
Intracristal space proteome mapping using super-resolution proximity labeling with isotope-coded probes
Abstract
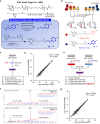
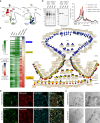
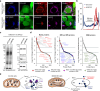
Proximity labeling with engineered ascorbate peroxidase (APEX) has been widely used to identify proteomes within various membrane-enclosed subcellular organelles. However, constructing protein distribution maps between two non-partitioned proximal spaces remains challenging with the current proximity labeling tools. Here, we introduce a proximity labeling approach using isotope-coded phenol probes for APEX labeling (ICAX) that enables the quantitative analysis of the spatial proteome at nanometer resolution between two distinctly localized APEX enzymes. Using this technique, we identify the spatial proteomic architecture of the mitochondrial intracristal space (ICS), which is not physically separated from the peripheral space. ICAX analysis further reveals unexpected dynamics of the mitochondrial spatiome under mitochondrial contact site and cristae organizing system (MICOS) complex inhibition and mitochondrial uncoupling, respectively. Overall, these findings highlight the importance of ICS for mitochondrial quality control under dynamic stress conditions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A patent application with M.-G.K. and H.W.R. has been filed by Seoul National University relating to this work. The remaining authors declare no competing interests.
Figures


Similar articles
-
4-plex quantitative glycoproteomics using glycan/protein-stable isotope labeling in cell culture.J Proteomics. 2025 Jan 6;310:105333. doi: 10.1016/j.jprot.2024.105333. Epub 2024 Oct 18. J Proteomics. 2025. PMID: 39426592
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Mitochondrial Contact Site and Cristae Organization System and F1FO-ATP Synthase Crosstalk Is a Fundamental Property of Mitochondrial Cristae.mSphere. 2021 Jun 30;6(3):e0032721. doi: 10.1128/mSphere.00327-21. Epub 2021 Jun 16. mSphere. 2021. PMID: 34133204 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

