A novel sequence-based transformer model architecture for integrating multi-omics data in preterm birth risk prediction
- PMID: 40835718
- PMCID: PMC12368083
- DOI: 10.1038/s41746-025-01942-2
A novel sequence-based transformer model architecture for integrating multi-omics data in preterm birth risk prediction
Abstract
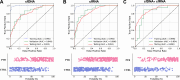
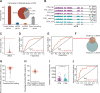
Preterm birth (PTB) significantly contributes to maternal and perinatal mortality and lifelong morbidity. While large language models (LLM) offer considerable potential for disease risk prediction and early detection, their application to PTB prediction using multi-omics data remains limited. We developed a novel transformer-based architecture for integrating cell free (cfDNA) and cfRNA sequencing data for PTB risk prediction. In the test set, the cfDNA LLM model achieved an AUC of 0.822, and the cfRNA LLM model achieved 0.851. Integrating cfDNA and cfRNA data within the transformer-based framework outperformed both, reaching an AUC of 0.890, a significant improvement over single-modality models. Additionally, we explored cfRNA and cfDNA integration using RNA editing and achieved an AUC of 0.82. This underscores the potential of multi-omics data fusion, with transformer-based architectures providing a powerful framework for disease risk assessment, and demonstrates the potential of AI-driven multi-omics for broader applications in precision obstetrics and biomedicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Automated Extraction of Mortality Information From Publicly Available Sources Using Large Language Models: Development and Evaluation Study.J Med Internet Res. 2025 Aug 18;27:e71113. doi: 10.2196/71113. J Med Internet Res. 2025. PMID: 40824124 Free PMC article.
-
Psychometric Evaluation of Large Language Model Embeddings for Personality Trait Prediction.J Med Internet Res. 2025 Jul 8;27:e75347. doi: 10.2196/75347. J Med Internet Res. 2025. PMID: 40627556 Free PMC article.
-
Differential Predictability of Preterm Birth Types: Strong Signals for Indicated Cases versus Limited Success in Spontaneous Preterm Birth.medRxiv [Preprint]. 2025 Jul 10:2025.07.09.25329712. doi: 10.1101/2025.07.09.25329712. medRxiv. 2025. PMID: 40672516 Free PMC article. Preprint.
-
Precision Neuro-Oncology in Glioblastoma: AI-Guided CRISPR Editing and Real-Time Multi-Omics for Genomic Brain Surgery.Int J Mol Sci. 2025 Jul 30;26(15):7364. doi: 10.3390/ijms26157364. Int J Mol Sci. 2025. PMID: 40806492 Free PMC article. Review.
-
Cervical pessary for preventing preterm birth in singleton pregnancies.Cochrane Database Syst Rev. 2022 Dec 1;12(12):CD014508. doi: 10.1002/14651858.CD014508. Cochrane Database Syst Rev. 2022. PMID: 36453699 Free PMC article.
References
Grants and funding
- JCYJ20240813144105007/Shenzhen Science and Technology Program
- 20240724152335001/Shenzhen Science and Technology Program
- LGKCYLWS2023019/Science and Technology Program of Longgang District, Shenzhen
- 82088102/National Natural Science Foundation of China
- 2019-I2M-5-064/CAMS Innovation Fund for Medical Sciences
LinkOut - more resources
Full Text Sources

