IWS1 positions downstream DNA to globally stimulate Pol II elongation
- PMID: 40835814
- PMCID: PMC12368070
- DOI: 10.1038/s41467-025-62913-5
IWS1 positions downstream DNA to globally stimulate Pol II elongation
Abstract
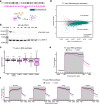
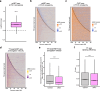
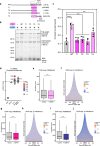
The protein IWS1 (Interacts with SPT6 1) is implicated in transcription-associated processes, but a direct role in RNA polymerase (Pol) II function is unknown. Here, we use multi-omics kinetic analysis after rapid depletion of IWS1 in human cells to show that loss of IWS1 results in a global decrease of RNA synthesis and a global reduction in Pol II elongation velocity. We then resolve the cryo-EM structure of the activated Pol II elongation complex with bound IWS1 and elongation factor ELOF1 and show that IWS1 acts as a scaffold and positions downstream DNA within the cleft of Pol II. In vitro assays show that the disordered C-terminal region of IWS1 that contacts the cleft of Pol II is responsible for stimulation of Pol II activity and is aided by ELOF1. Finally, we find that the defect in transcription upon IWS1 depletion leads to a decrease of histone H3 tri-methylation at residue lysine-36 (H3K36me3), but that this secondary effect is an indirect function of IWS1. In summary, our structure-function analysis establishes IWS1 as a Pol II-associated elongation factor that acts globally to stimulate Pol II elongation velocity and ensure proper co-transcriptional histone methylation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Roeder, R. G. & Rutter, W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature224, 234–237 (1969). - PubMed
-
- Sentenac, A. Eukaryotic RNA polymerases. CRC. Crit. Rev. Biochem18, 31–90 (1985). - PubMed
-
- Cramer, P. Eukaryotic transcription turns 50. Cell179, 808–812 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

