Targeted CRISPR screens reveal genes essential for Cryptosporidium survival in the host intestine
- PMID: 40835841
- PMCID: PMC12368253
- DOI: 10.1038/s41467-025-63012-1
Targeted CRISPR screens reveal genes essential for Cryptosporidium survival in the host intestine
Abstract
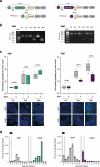
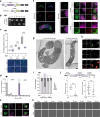
The Cryptosporidium parasite is one of the leading causes of diarrheal morbidity and mortality in children, and adolescent infections are associated with chronic malnutrition. There are no vaccines available for protection and only one drug approved for treatment that has limited efficacy. A major barrier to developing new therapeutics is a lack of foundational knowledge of Cryptosporidium biology, including which parasite genes are essential for survival and virulence. Here, we iteratively improve the tools for genetically manipulating Cryptosporidium and develop a targeted CRISPR-based screening method to rapidly assess how the loss of individual parasite genes influence survival in vivo. Using this method, we examine the parasite's pyrimidine salvage pathway and a set of leading Cryptosporidium vaccine candidates. From this latter group, using inducible knockout, we determined the parasite gene known as Cp23 to be essential for survival in vivo. Parasites deficient in Cp23 were able to replicate within and emerge from infected epithelial cells, yet unable to initiate gliding motility which is required for the reinfection of neighbouring cells. The targeted screening method presented here is highly versatile and will enable researchers to more rapidly expand the knowledge base for Cryptosporidium infection biology, paving the way for new therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet382, 209–222 (2013). - PubMed
-
- Kotloff, K. L. et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Glob. Health7, e568–e584 (2019). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

