Integrated genome mining and molecular networking uncover the biosynthetic potential of a novel mangrove-derived strain Streptomyces sp. B1866
- PMID: 40835898
- PMCID: PMC12366066
- DOI: 10.1186/s12864-025-11966-3
Integrated genome mining and molecular networking uncover the biosynthetic potential of a novel mangrove-derived strain Streptomyces sp. B1866
Abstract
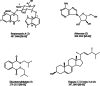
Mangrove-derived actinomycetes represent a prolific reservoir of natural products, characterized by diverse chemical structures and significant pharmacological properties. Here, a mangrove-derived Streptomyces sp. B1866 was proposed to represent a novel species within the genus Streptomyces due to the low 16S rRNA gene sequence similarity (< 97%), and low overall genome relatedness indices (dDDH, 24.1%-24.7%; ANI, 80.1%-80.6%) with its closely related species. A whole genome sequence of strain B1866 comprised 42 secondary metabolite-biosynthetic gene clusters (BGCs), more than half of which exhibited low similarity (< 70%) to characterized BGCs, showing a potential for the production of compounds that are either uncharacterized or not associated with known structures. UPLC-MS/MS-based molecular networking further revealed the biosynthetic potential of strain B1866 since several nodes could not be assigned to the previously reported compounds. Eventually, investigation chemical constitutes in the fermentation of strain B1866 led to the isolation of a previously undescribed benzoxazole, designated streptoxazole A (1), along with 3 known compounds adenosine (2), diisobutyl phthalate (3), and ergosta-5,7,22-trien-3-ol (4). Streptoxazole A (1) possessed anti-inflammatory properties, inhibiting LPS-induced nitric oxide (NO) production with IC50 values of 38.4 μM. This study exemplifies the discovery of novel molecules from mangrove-derived Streptomyces species.
Background Mangrove-derived microorganisms have been confirmed to produce secondary metabolites with various chemical structures and pharmaceutical activity. By integrating genome mining with UPLC-MS/MS based molecular networking approach, the metabolic potential of a novel mangrove-derived Streptomyces species B1866 was confirmed. Further investigation of the metabolite profile of strain B1866 led to the isolation of three known secondary metabolites, as well as a novel benzoxazole compound with anti-inflammatory property.
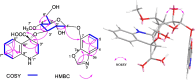
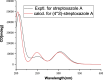
Results In this study, a Streptomyces strain B1866 isolated from mangrove sediments was identified and characterized. The whole genome of strain B1866 was sequenced to identify 21 BGCs involved in the biosynthesis of polyketides (PKS), non-ribosomal peptides (NRPS), and PKS-NRPS hybrid metabolites, highlighting its metabolic potential. Meanwhile, UPLC-MS/MS-based molecular networking revealed the presence of products with diverse structures in the crude extracts of strain B1866. Subsequently, compound 1, namely streptoxazole A (1), with a rare benzoxazole moiety was isolated. The structure of 1 was deduced from the analysis of 1D/2D NMR, HRESIMS data, and electronic circular dichroism (ECD) comparisons. The biosynthetic pathway of 1 required the shikimate pathway to generate precursor 3-amino-1,2-benzenediol and 2-(2-carboxyacetyl) benzoic acid. The isolated compound streptoxazole A (1) showed potent inhibitory effect toward NO production in LPS-induced RAW264.7 cells (IC50 38.4 μM).
Conclusions In conclusion, a novel benzoxazole, designated streptoxazole A (1), with anti-inflammatory property was isolated from a novel mangrove-derived Streptomyces sp. B1866. The integration of genome mining and UPLC-MS/MS-based molecular networking not only unveiled the metabolic potential of strain B1866 but also provided evidence for the discovery of novel chemicals from mangrove-derived Streptomyces species.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12864-025-11966-3.
Keywords: Streptomyces sp.; Biosynthetic potential; Genome mining; Molecular networking; Novel benzoxazole.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Primavera JH, Friess DA, Van Lavieren H, Lee SY. The Mangrove Ecosystem. World Seas: An Environmental Evaluation (second edition), 2019; 1–34. 10.1016/b978-0-12-805052-1.00001-2.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

