Genetic and molecular landscape of comorbidities in people living with HIV
- PMID: 40835971
- PMCID: PMC12532687
- DOI: 10.1038/s41591-025-03887-1
Genetic and molecular landscape of comorbidities in people living with HIV
Abstract
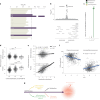
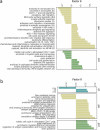
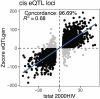
People living with HIV (PLHIV) have an increased susceptibility to non-AIDS comorbidities. In this study, we systematically profiled 1,342 PLHIV across five omics layers and immune function. We found latent factors, resulting from integrating epigenomics, transcriptomics, proteomics, metabolomics and immune responses, linked to cardiovascular diseases, the presence of carotid plaque and chronic obstructive pulmonary disease in PLHIV. Mapping four omics layers to genetic variation identified 5,962 molecular quantitative trait loci, illustrating a common genetic regulation in PLHIV compared to healthy individuals. By performing Mendelian randomization, we uncovered host genetic-driven changes in baseline molecules causally related to immune responses upon stimulation with inactivated pathogens. Lastly, we uncovered that the inflammasome, genetically regulated by the NLRP12 locus, contributes to systemic inflammation across multiple molecular layers. This study offers a unique catalog of genetic and molecular determinants of immune function in PLHIV and elucidates molecular pathways driving inter-individual variation in immune response and comorbidities.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






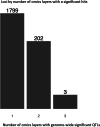


References
-
- Fauci, A. S. & Folkers, G. K. Toward an AIDS-free generation. JAMA308, 343–344 (2012). - PubMed
MeSH terms
Substances
Grants and funding
- 390874280/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 497673685/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 833247/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Medical

