Induced somatic mutation accumulation during skeletal muscle regeneration reduces muscle strength
- PMID: 40836125
- PMCID: PMC12443595
- DOI: 10.1038/s43587-025-00941-y
Induced somatic mutation accumulation during skeletal muscle regeneration reduces muscle strength
Abstract
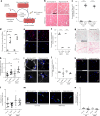
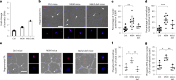
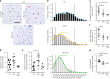
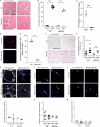
Aging is associated with a progressive decline in tissue function and regenerative capacity, partly due to genomic instability, one of the hallmarks of aging1,2. Genomic instability encompasses DNA damage and the accumulation of somatic mutations in post-zygotic cells, yet the specific impact of these mutations on age-related tissue dysfunction remains poorly understood. To address this, we developed a mouse model in which genomic instability was induced specifically in muscle progenitor cells3 through targeted deletion of the Msh2 (ref. 4) and Blm5 genes. This allowed us to assess how elevated DNA damage and somatic mutations, from single-nucleotide variants (SNVs) to structural variants, affect muscle regeneration following injury. These mice exhibited impaired muscle regeneration, characterized by smaller muscle fibers, reduced muscle mass gain and decreased grip strength. Importantly, similar muscle deficits were observed in a second mouse model where somatic mutations were elevated with less substantial DNA damage. These findings provide evidence that the accumulation of somatic mutations can potentially compromise the function of somatic cells, contributing to the aging phenotype in skeletal muscle.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell186, 243–278 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

