Functional characterization of AsGRAS24 in Avena sativa reveals its role in enhancing salt tolerance via antioxidant and hormonal regulation in transgenic Arabidopsis thaliana
- PMID: 40836209
- PMCID: PMC12366120
- DOI: 10.1186/s12870-025-07061-0
Functional characterization of AsGRAS24 in Avena sativa reveals its role in enhancing salt tolerance via antioxidant and hormonal regulation in transgenic Arabidopsis thaliana
Abstract
Background: GRAS transcription factors are a type of transcription factor found only in plants. Among these, PAT1 subfamily proteins are known to help plants resist abiotic stress.
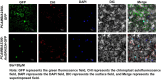
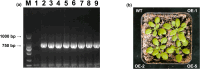
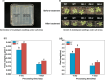
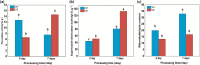
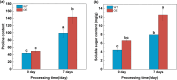
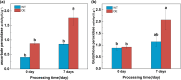
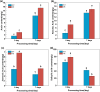
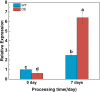
Results: To study the GRAS family genes in oat and check if GRAS transcription factors are linked to salt tolerance, this research cloned the AsGRAS24 gene using oat leaf cDNA. The gene was analyzed using bioinformatics tools, and its cellular localization was determined. Through the Agrobacterium-mediated genetic transformation method, the target gene was introduced into the model plant Arabidopsis thaliana, and then the functional mechanism and regulatory network of this gene in plant salt stress tolerance were systematically studied. The results showed that the AsGRAS24 gene produces a protein with 291 amino acids, weighing 32.44 kDa, and has a pI value of 7.1. The protein was found in the nucleus. The AsGRAS24 protein shows the highest sequence similarity to a specific gene in OsGRAS24 (Oryza sativa). The study also compared wild-type A. thaliana seeds (WT) and transgenic AsGRAS24 A. thaliana plants under salt stress. It was found that the AsGRAS24 gene made the transgenic plants more salt-tolerant. As the salt stress continued, the transgenic plants exhibited higher levels of antioxidant enzymes, osmotic adjustment substances, hormones, and gene expression compared to the wild-type (WT) plants (P < 0.05). The results suggest that AsGRAS24 is involved in helping transgenic A. thaliana tolerate salt. Therefore, the AsGRAS24 gene is essential in enhancing salt tolerance in transgenic A. thaliana.
Conclusions: This research helps in understanding how oats resist stress at the molecular level, providing a reference for further study of its function in the development and stress resistance of Gramineae.
Keywords: Avena sativa; Bioinformatics analysis; Phenotype; Physiological index; Salt tolerance; Transcription factors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Overexpression of a Malus baccata (L.) Borkh WRKY Factor Gene MbWRKY33 Increased High Salinity Stress Tolerance in Arabidopsis thaliana.Int J Mol Sci. 2025 Jun 18;26(12):5833. doi: 10.3390/ijms26125833. Int J Mol Sci. 2025. PMID: 40565296 Free PMC article.
-
Overexpression of SiGSTU24 enhances salt tolerance in transgenic Arabidopsis.BMC Plant Biol. 2025 Jul 30;25(1):986. doi: 10.1186/s12870-025-07021-8. BMC Plant Biol. 2025. PMID: 40739479 Free PMC article.
-
The transcription factor ThDOF8 binds to a novel cis-element and mediates molecular responses to salt stress in Tamarix hispida.J Exp Bot. 2024 May 20;75(10):3171-3187. doi: 10.1093/jxb/erae070. J Exp Bot. 2024. PMID: 38400756
-
Identification of crucial genes sustaining the curled leaf phenotype in perennial transgenic poplar via advanced proteomic and phosphoproteomic analyses.J Proteomics. 2025 Aug 15;319:105471. doi: 10.1016/j.jprot.2025.105471. Epub 2025 Jun 3. J Proteomics. 2025. PMID: 40472892 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Zhang JZ. Overexpression analysis of plant transcription factors[J]. Curr Opin Plant Biol. 2003;6(5):430–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

