Cold stress enhances cryotolerance in Lacticaseibacillus rhamnosus B6 via membrane lipid remodeling and differential protein expression
- PMID: 40837524
- PMCID: PMC12363473
- DOI: 10.1016/j.crmicr.2025.100453
Cold stress enhances cryotolerance in Lacticaseibacillus rhamnosus B6 via membrane lipid remodeling and differential protein expression
Abstract
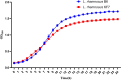
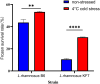
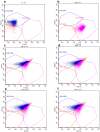
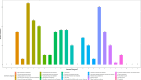
Understanding the molecular mechanisms underlying the cryotolerance of lactic acid bacteria is critical for preserving their viability in food processing. In this study, Lacticaseibacillus rhamnosus B6 and KF7 with different phenotypic properties were pretreated at 4 °C for 2 h before liquid nitrogen freezing. Cold-stressed B6 exhibited significantly higher survival (53 %) than KF7 (30 %) and untreated controls (44 % vs. 10 %, p < 0.05), with few disrupted cells observed under SEM. Cold stress altered the membrane fluidity in B6 by increasing unsaturated fatty acids (UFA) and the ratio of UFA to saturated fatty acids (UFA/SFA) from 1.36 to 1.62 (p < 0.05). 219 proteins, primarily those involved in fatty acid biosynthesis, translation and transport processes were up regulated in cold-stressed B6, compared to untreated counterparts. Noteworthy, capsules of B6 likely contributed to its higher cryotolerance. Our findings revealed a dual cryotolerance mechanism in L. rhamnosus B6, i.e., dynamic lipidome remodeling and coordinated protein expression regulation. This study highlighted the importance of the cell surface features of individual strain of L. rhamnosus and cold stress treatment in preparing probiotics as well as direct to vat starter cultures with high cell vitality employing deep freezing process.
Keywords: Cold stress; Cryotolerance; Fatty acids; Lacticaseibacillus; Proteomics.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Ayu Eka Pitaloka D., Izzati A., Rafa Amirah S., Abdan Syakuran L., Muhammad Irham L., Darumas Putri A., Adikusuma W. Bioinformatics analysis to uncover the potential drug targets responsible for mycobacterium tuberculosis peptidoglycan and lysine biosynthesis. Bioinform. Biol. Insights. 2023;17 doi: 10.1177/11779322231171774. - DOI - PMC - PubMed
-
- Bustos A.Y., Taranto M.P., Gerez C.L., Agriopoulou S., Smaoui S., Varzakas T., Enshasy H.A.E. Recent advances in the understanding of stress resistance mechanisms in probiotics: relevance for the design of functional food systems. Probiotics. Antimicrob. Proteins. 2024 doi: 10.1007/s12602-024-10273-9. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

