Comparative transcriptomics analysis of the Oleidesulfovibrio alaskensis G20 biofilms grown on copper and polycarbonate surfaces
- PMID: 40837593
- PMCID: PMC12363597
- DOI: 10.1016/j.bioflm.2025.100309
Comparative transcriptomics analysis of the Oleidesulfovibrio alaskensis G20 biofilms grown on copper and polycarbonate surfaces
Abstract
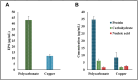
Sulfate-reducing bacterial (SRB) biofilms are prevalent across diverse environments, playing key roles in biogeochemical sulfur cycling while also contributing to industrial challenges such as biofouling and biocorrosion. Understanding the genetic and physiological adaptations of SRB biofilms to different surfaces is crucial for developing mitigation strategies. This study presents a comparative transcriptomic analysis of Oleidesulfovibrio alaskensis G20 biofilms grown on copper and polycarbonate surfaces, aimed at elucidating their differential responses at the molecular level. RNA sequencing revealed 1255 differentially expressed genes, with copper-grown biofilms exhibiting upregulation of Dde_1570 (flagellin; log2FC 2.31) and Dde_0831 (polysaccharide chain length determinant; log2FC 1.15), highlighting enhanced motility and extracellular polymeric substance production. Conversely, downregulated genes on copper included Dde_0132 (Cu/Zn efflux transporter; log2FC -3.37) and Dde_0369 (methyl-accepting chemotaxis protein; log2FC -1.19), indicating a metabolic shift and stress adaptation to metal exposure. Morphological analysis via SEM revealed denser biofilm clusters with precipitates on copper, whereas biofilms on polycarbonate were more dispersed. AFM analysis showed a 4.6-fold increase in roughness on copper (44.3 ± 3.1 to 205.89 ± 8.7 nm) and a 3.8-fold increase on polycarbonate (521.12 ± 15.2 to 1975.64 ± 52.6 nm), indicating surface erosion and structural modifications. Protein-protein interaction analysis identified tightly regulated clusters associated with ribosomal synthesis, folate metabolism, and quorum sensing, underscoring their role in biofilm resilience. Additionally, functional annotations of uncharacterized genes revealed potential biofilm regulators, such as Dde_4025 (cytochrome-like protein; log2FC 4.18) and Dde_3288 (DMT superfamily permease; log2FC 3.55). These findings provide mechanistic insights into surface-dependent biofilm formation, with implications for designing antifouling materials and controlling microbial-induced corrosion.
Keywords: Biocorrosion; Biofilm; Metallic; Non-metallic; SRB; Surface roughness; Transcriptomics.
© 2025 Published by Elsevier B.V.
Conflict of interest statement
I have nothing to declare.
Figures









Similar articles
-
Copper-coated carbon-infiltrated carbon nanotube surfaces effectively inhibit Staphylococcus aureus and Pseudomonas aeruginosa biofilm formation.Appl Environ Microbiol. 2025 Aug 20;91(8):e0105325. doi: 10.1128/aem.01053-25. Epub 2025 Jul 8. Appl Environ Microbiol. 2025. PMID: 40626868 Free PMC article.
-
Sulfate-reducing bacteria: Unraveling biofilm complexity, stress adaptation, and strategies for corrosion control.Sci Total Environ. 2025 Aug 16;998:180226. doi: 10.1016/j.scitotenv.2025.180226. Online ahead of print. Sci Total Environ. 2025. PMID: 40819394 Review.
-
Molecular regulation of conditioning film formation and quorum quenching in sulfate reducing bacteria.Front Microbiol. 2022 Oct 31;13:1008536. doi: 10.3389/fmicb.2022.1008536. eCollection 2022. Front Microbiol. 2022. PMID: 36386676 Free PMC article. Review.
-
Meta-Transcriptomic Response to Copper Corrosion in Drinking Water Biofilms.Microorganisms. 2025 Jun 30;13(7):1528. doi: 10.3390/microorganisms13071528. Microorganisms. 2025. PMID: 40732036 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
References
-
- Krishnan S., Patil S.A., Nancharaiah Y. Material-microbes interactions. Elsevier; 2023. Environmental microbial biofilms: formation, characteristics, and biotechnological applications; pp. 3–45.
-
- Shineh G., et al. Biofilm formation, and related impacts on healthcare, food processing and packaging, industrial manufacturing, marine industries, and sanitation–a review. Appl Microbiol. 2023;3(3):629–665.
-
- Tuck B., et al. A critical review of marine biofilms on metallic materials. npj Mater Degrad. 2022;6(1):25.
LinkOut - more resources
Full Text Sources
Miscellaneous

