Targets and mechanisms of epigenetic regulation in the temperate cereal vernalisation process
- PMID: 40838075
- PMCID: PMC12361225
- DOI: 10.3389/fpls.2025.1520593
Targets and mechanisms of epigenetic regulation in the temperate cereal vernalisation process
Abstract
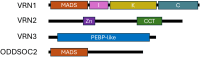
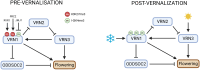
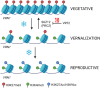
Vernalisation is a prolonged cold exposure that synchronises flowering with favourable seasonal conditions, protecting reproductive development from winter stress and optimising crop yields. By 'recording' winter, this biological process displays memory and becomes progressively more difficult to reverse. In the well-studied model Arabidopsis, vernalisation epigenetically silences the gene locus of the floral repressor FLC. Temperate cereals, including crops such as wheat and barley, respond to a similar vernalisation cue with a memory property and recent functional studies also support an epigenetic mechanism. Current evidence points to the flowering promoter VRN1 as the primary site for storing this memory. Because vernalisation in cereals appears to rely on epigenetic activation of VRN1, rather than repression as in Arabidopsis, the specific histone marks responsible for storing this epigenetic memory are possibly different. This highlights the need for further research to identify the specific genes and histone modifications involved, and to fully elucidate the mechanisms underlying vernalisation memory in cereals. The goal of this review is to synthesise recent advances in our understanding of the epigenetic regulation of vernalisation in temperate cereals. Therefore, this review focuses on the roles of key genes such as VRN1, VRN3, and ODDSOC2, and examines the dynamic chromatin landscape associated with vernalisation-induced flowering. In particular, we investigate the possible interplay of chromatin marks involved in the epigenetic activation of VRN1. By synthesising current knowledge and highlighting unresolved questions, this review aims to provide a framework for future research in the field of cereal vernalisation memory.
Keywords: VRN1; devernalisation; histone modification; temperate cereals; vernalisation.
Copyright © 2025 Daneels, Martinez-Barrales, Bosmans and Geuten.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Technological aids for the rehabilitation of memory and executive functioning in children and adolescents with acquired brain injury.Cochrane Database Syst Rev. 2016 Jul 1;7(7):CD011020. doi: 10.1002/14651858.CD011020.pub2. Cochrane Database Syst Rev. 2016. PMID: 27364851 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

