Beyond the genome: the role of functional markers in contemporary plant breeding
- PMID: 40838076
- PMCID: PMC12361171
- DOI: 10.3389/fpls.2025.1637299
Beyond the genome: the role of functional markers in contemporary plant breeding
Abstract
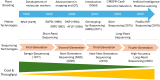
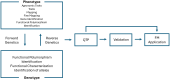
Functional markers (FMs) are derived from polymorphisms that confer phenotypic trait variation, making them powerful tools in plant breeding. Unlike random markers, for which trait associations are unknown, or at best established via linkage or quantitative trait locus (QTL) analysis, FMs are associated with causative polymorphisms, providing precise and reliable information for trait selection. Since the concept of FMs was first proposed in 2003, the emergence and adoption of technologies that were not available at the time have significantly advanced FM discovery and application by enhancing the ability to precisely identify causal variants underlying complex traits, which is a critical prerequisite for FM development. Novel technologies such as high-throughput sequencing, multi-omics, gene editing, and advanced computational tools have enabled the precise identification and functional validation of DNA polymorphisms associated with trait variation. FMs can be used in genomic selection (GS) and modern plant breeding programs by improving selection efficiency and accuracy. While FMs provide numerous benefits, challenges still remain regarding their stability and transferability, and innovative approaches to overcome these limitations are continually being explored. The role of FMs in plant breeding is expected to grow as functional annotation of genomes improves and technologies like genome editing become more accessible. These developments will enable breeders to effectively integrate FMs into breeding pipelines for accelerating genetic gains and addressing global agricultural challenges.
Keywords: functional markers (FMs); genomic selection (GS); polymorphism; random markers (RDMs); sequencing.
Copyright © 2025 Park, Silva, Lübberstedt and Scott.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Acevedo-Garcia J., Spencer D., Thieron H., Reinstädler A., Hammond-Kosack K., Phillips A. L., et al. (2017). mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 15, 367–378. doi: 10.1111/pbi.12631, PMID: - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

