Enhancing wheat genomic prediction by a hybrid kernel approach
- PMID: 40838078
- PMCID: PMC12363290
- DOI: 10.3389/fpls.2025.1605202
Enhancing wheat genomic prediction by a hybrid kernel approach
Abstract
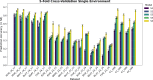
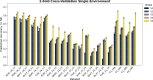
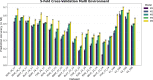
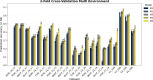
This study integrates genomic and pedigree data by leveraging advanced modeling techniques, aiming to enhance the predictive performance of genomic selection models by capturing complex genetic relationships through the interaction of both matrices and exploring the utility of non-linear methods, such as kernel matrices. Our goal was to improve genomic prediction accuracy by combining the pedigree-based or genetic similarity matrix ( A ) with the genomic similarity matrix ( G ). Using various wheat datasets, we performed five single-environment models and five multi-environment models that incorporated genotype-by-environment (G × E) interactions. The proposed models S5 and M5 significantly enhanced prediction accuracy by incorporating two novel symmetric kernels, C and P , derived from the interaction of genomic and pedigree matrices. These hybrid kernels captured additional, independent genetic variation not explained by conventional matrices. The proposed prediction model outperformed the standard conventional models in most single-environment and multi-environment models. The genomic models with non-linear kernels were better predictors than the linear prediction models.
Keywords: genomics; genotype by environment interaction; merging genomics and pedigree; pedigree; single-environment.
Copyright © 2025 Cuevas, Crossa, Montesinos-López, Martini, Gerard, Ortegón, Dreisigacker, Govindan, Pérez-Rodríguez, Saint Pierre, Herrera, Montesinos-López and Vitale.
Conflict of interest statement
Author JM was employed by the company Aardevo B.V. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

