Spatio-temporal processes in autophagosome-lysosome fusion
- PMID: 40838105
- PMCID: PMC12362064
- DOI: 10.1515/mr-2024-0095
Spatio-temporal processes in autophagosome-lysosome fusion
Abstract
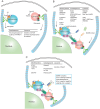
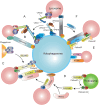
Macroautophagy/autophagy is a lysosome-dependent degradation process involved in cellular energy metabolism, recycling and quality control. Autophagy is a highly dynamic and precisely regulated process, which contains four major steps: autophagic membrane initiation and cargo recognition, autophagosome formation, autophagosome-lysosome fusion and lysosomal degradation. During the terminal phase of autophagy, the merging of the autophagosome and lysosome membranes is critical for the effective breakdown of sequestered cargoes. However, the participated molecules and the interplay among them have not been fully uncovered. The spatiotemporal property of these molecules is crucial for maintaining the orderly fusion of autophagosomes and lysosomes, otherwise it may lead to fusion disorders. In this article, we tend to summarize the molecules mediating autophagosome-lysosome fusion into two categories: effector molecules and regulatory molecules. The effector molecules are soluble N-ethylmaleimide-sensitive factor attachment protein receptor and tethering proteins, and the latter category contains phosphatidylinositol, Rab GTPases and ATG8-family proteins. The spatio-temporal properties of these autophagosome-lysosome fusion mediating molecules will be featured in this review.
Keywords: autophagosome-lysosome fusion; autophagy; sensitive factor attachment protein receptor; syntaxin 17.
© 2025 the author(s), published by De Gruyter, Berlin/Boston.
Conflict of interest statement
Conflict of interest: The authors state no conflict of interest.
Figures


Similar articles
-
An initial HOPS-mediated fusion event is critical for autophagosome transport initiation from the axon terminal.Autophagy. 2024 Oct;20(10):2275-2296. doi: 10.1080/15548627.2024.2366122. Epub 2024 Jun 20. Autophagy. 2024. PMID: 38899385 Free PMC article.
-
Comprehensive knockout analysis of the RAB family small GTPases reveals an overlapping role of RAB2 and RAB14 in autophagosome maturation.Autophagy. 2025 Jan;21(1):21-36. doi: 10.1080/15548627.2024.2374699. Epub 2024 Jul 10. Autophagy. 2025. PMID: 38953305 Free PMC article.
-
Molecular Mechanism of Autophagosome-Lysosome Fusion in Mammalian Cells.Cells. 2024 Mar 13;13(6):500. doi: 10.3390/cells13060500. Cells. 2024. PMID: 38534345 Free PMC article. Review.
-
Blocking autophagosome closure manifests the roles of mammalian Atg8-family proteins in phagophore formation and expansion during nutrient starvation.Autophagy. 2025 May;21(5):1059-1074. doi: 10.1080/15548627.2024.2443300. Epub 2025 Jan 20. Autophagy. 2025. PMID: 39694607 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
