Perioperative IDH inhibition in treatment-naive IDH-mutant glioma: a pilot trial
- PMID: 40841487
- PMCID: PMC12532589
- DOI: 10.1038/s41591-025-03884-4
Perioperative IDH inhibition in treatment-naive IDH-mutant glioma: a pilot trial
Abstract
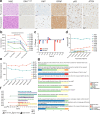
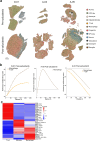
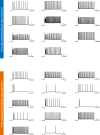
Mutant isocitrate dehydrogenase (mIDH) inhibition significantly improves progression-free survival in patients with mIDH WHO grade 2 glioma; however, a large proportion of patients will progress, and mechanisms of adaptation to mIDH inhibition remain poorly understood. Perioperative studies with evaluation of paired pre- and post-treatment samples enable detailed understanding of drug response, facilitating biomarker development, but are rare in glioma owing to safety and cost concerns. Here we conducted a single-arm, open-label feasibility perioperative trial in patients with mIDH1 low-grade glioma, treatment naive to radiation and chemotherapy, with safusidenib (AB-218/DS-1001b), an orally available small-molecule inhibitor of mIDH1. As of 8 November 2024, 10 patients were enrolled and have completed the perioperative component, with a median follow-up of 14 months. Patients continue postoperative safusidenib with ongoing follow-up for safety and efficacy. The primary endpoint showed the feasibility and acceptability of conducting a two-stage perioperative trial. One patient experienced a serious surgery-related adverse event, and ten reported safusidenib-related adverse events; most were grade 1, and one experienced grade 3 elevation of transaminases. Tumor 2-hydroxyglutarate quantification revealed on-target activity, associated with alterations in differentiation programs and neural excitability, functionally validated in post hoc analysis by patch-clamp electrophysiology. Taken together, these results provide a detailed investigation of observations associated with mIDH inhibition in glioma. The study shows the safety and feasibility of this perioperative approach, which can be applied broadly in clinical trial design, serving as proof of concept for advancing drug development in glioma. ClinicalTrials.gov registration: NCT05577416 .
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.J.D. reports research funding from AnHeart Therapeutics to the institute (Royal Melbourne Hospital). J.R.W. reports research funding from AnHeart Therapeutics to the institute (WEHI), receiving consulting fees from AnHeart Therapeutics and Servier, being on advisory boards for Roche and Merck, and being a data safety monitoring member for Telix Pharmaceuticals. The other authors declare no competing interests.
Figures











References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

