Programmed cell revival from imminent cell death enhances tissue repair and regeneration
- PMID: 40841709
- PMCID: PMC12489119
- DOI: 10.1038/s44318-025-00540-y
Programmed cell revival from imminent cell death enhances tissue repair and regeneration
Abstract
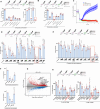
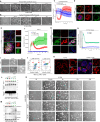
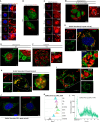
Cell recovery from near-death states is a critical yet poorly understood aspect of cell biology. Here, we describe a tightly-regulated programmed cell revival process after exposure of cells to cell death-inducing lysosomotropic agents, such as L-leucyl-L-leucine methyl ester (LLOMe). In the initial stage of cell recovery, we observe increased chromatin accessibility and upregulation of genes and pathways associated with embryonic development, regeneration, stemness, and inflammation. Subsequently, vital pathways governing metabolism, organelle biogenesis, membrane trafficking, transport, and cytoskeleton remodeling are activated, resulting in the complete renewal of cells. Consistent with the links of this transcriptional profile to tissue repair and regeneration, we found LLOMe to enhance the healing of skin wounds and corneal alkali burns in mice, promote hematopoietic progenitor/stem cell production in Drosophila melanogaster, induce tadpole tail regeneration in frogs, and mediate axon regeneration in Caenorhabditis elegans. Using both genetic and pharmacological approaches, we show NF-ĸB signaling to be critical for both cell revival and regeneration. This study characterizes cell revival from near-death conditions as a programmed cell-intrinsic mechanism, which could be harnessed for therapeutic applications in regenerative medicine.
Keywords: Anastasis; LLOMe; Lysosomal Cell Death and Revival; NF-ĸB Signaling in Cell Revival and Regeneration; Programmed Cell Revival.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures











References
-
- Alberts B, Johnson AD, Morgan D, Roberts K, Walter P, Raff MC, Lewis J (2014) Molecular biology of the cell. Garland Science, Taylor and Francis Group
MeSH terms
Substances
Grants and funding
- CRG/2022/002777/ANRF
- NCP070301/Council of Scientific and Industrial Research, India (CSIR)
- DHR-GIA/2020-NCD-1/Indian Council of Medical Research (ICMR)
- PCT/IN2025/051017/PCT
- IIRP-2023-1988/Indian Council of Medical Research (ICMR)
- BT/PR45223/MED/29/1613/2022/Department of Biotechnology, Ministry of Science and Technology, India (DBT)
- BT/PR23942/BRB/10/1808/2019/Department of Biotechnology, Ministry of Science and Technology, India (DBT)
- IA/S/22/1/506243/Wellcome Trust DBT India Alliance (India Alliance)
- IIRPIG-2023-0814/Indian Council of Medical Research (ICMR)
- WT_/Wellcome Trust/United Kingdom
- BT/PR41956/MED/97/515/2021/Department of Biotechnology, Ministry of Science and Technology, India (DBT)
- ICMRCAREP-2023-0000176/Indian Council of Medical Research (ICMR)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

