Subspecific variation in gut microbiota of North American bison in a sympatric setting reveals differentially abundant taxa
- PMID: 40841983
- PMCID: PMC12372302
- DOI: 10.1186/s42523-025-00451-7
Subspecific variation in gut microbiota of North American bison in a sympatric setting reveals differentially abundant taxa
Abstract
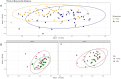
Gut microbiomes play critical roles in host-environment interactions, reflecting habitat and foraging niches. North American bison (Bison bison) subspecies—plains bison (B. bison bison) and wood bison (B. bison athabascae)—exhibit limited genetic variation from historic population bottleneck events, potentially undermining their evolutionary potential. Understanding variation in gut microbiota composition between subspecies may shed light on genetic, phenotypic, and ecological divergence relevant to their adaptive capacities. Using 16S rRNA metabarcoding of fecal samples, we characterized the gut microbiota of both subspecies in the sympatric environment of Elk Island National Park, providing insight into potential phylogenetic gut microbiome divergence. Like other ruminants, the gut microbial community of both subspecies consists primarily of the bacterial phyla Firmicutes and Bacteroidetes. Subspecific classification explained no significant differences in alpha diversity (p > 0.05) in the overall dataset, but has a potentially significant effect on beta diversity (p < 0.05, R2 = 0.04). Gut microbiota divergence between subspecies may be driven by differential abundance of specific taxa and associated functional pathways, likely influenced by dietary preferences, ancestral phenotypes, and historical ranges. Our findings support further investigation into diet-microbiome relationships between subspecies in sympatric environments and metagenomic approaches to explore functional differences in the gut microbiome.
Supplementary Information: The online version contains supplementary material available at 10.1186/s42523-025-00451-7.
Keywords: Gut microbiome; North American bison; Phylogenetic divergence; Subspecific variation.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The authors of this study confirm that we have adhered to all prevailing local, national and international regulations and conventions, and have followed normal ethical scientific practices throughout the study. Sampling was performed under Parks Canada Research and Collection Permit EINP-2023-45630. Competing interests: The authors declare no competing interests.
Figures


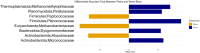


References
Grants and funding
LinkOut - more resources
Full Text Sources
