PRRSV evades innate immune cGAS-STING antiviral function via its Nsp5 to deter STING translocation and activation
- PMID: 40842188
- PMCID: PMC12377093
- DOI: 10.1080/21505594.2025.2548625
PRRSV evades innate immune cGAS-STING antiviral function via its Nsp5 to deter STING translocation and activation
Abstract
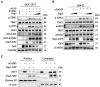
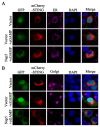
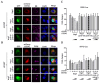
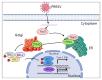
Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) is an important pathogen that seriously endangers pig breeding, causing significant economic losses to the global swine industry. Our previous study found that the DNA sensing innate cGAS-STING signaling pathway plays an important role in inducing interferon (IFN) upon PRRSV infection and inhibition of PRRSV replication. However, the mechanism underlying immune evasion by PRRSV remains unclear. In the current study, we found that PRRSV non-structural protein 5 (Nsp5) strongly inhibits the cGAS-STING-IFN antiviral response. Furthermore, we found that Nsp5 interacts with STING, blocking STING transport from the ER to the Golgi apparatus and interfering with STING recruitment of TBK1/IKKε/IRF3. Finally, we demonstrated that the Nsp5 36-47 and 58-67 amino acid regions are critical for inhibiting STING activity and PRRSV replication. This study describes a novel mechanism by which PRRSV suppresses the host innate antiviral response and has implications for our understanding of PRRSV pathogenesis.
Keywords: Nsp5; PRRSV; STING; immune evasion.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
