Systematic modulation of bacterial resource allocation by perturbing RNA polymerase availability via synthetic transcriptional switches
- PMID: 40842238
- PMCID: PMC12370630
- DOI: 10.1093/nar/gkaf814
Systematic modulation of bacterial resource allocation by perturbing RNA polymerase availability via synthetic transcriptional switches
Abstract
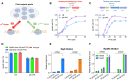
Gene regulation and its interplay with physiological behaviors are the central topics of modern biology. Classical studies on gene regulation focus intensively on specific regulatory mechanisms of transcription. Nevertheless, the genome-wide impact of RNA polymerase (RNAP) availability on gene expression remains poorly understood. Here we developed two synthetic transcriptional switches to systematically titrate the expression of either ${\sigma ^A}$ (SigA, housekeeping sigma factor) or RpoBC (core enzyme) in Bacillus subtilis. Both systems effectively modulated cell growth, but with fundamentally distinct mechanisms. SigA limitation triggered significant resource reallocation, redirecting cellular investment from biosynthetic pathways to alternative cellular pathways, which could further facilitate the engineering of dynamic growth-bioproduction switch. In contrast, RpoBC depletion caused only weak changes of gene expression but induced ribosomal inactivation through blocking translation initiation. Notably, RpoBC depletion induced DNA damage response and increased the DNA damage sensitivity of bacteria, suggesting transcription-coupled repair as a critical survival mechanism. Our findings delineate two regulatory paradigms of resource allocation that are associated with the interplay between RNAP availability and bacterial physiological state, "abundance-based" and "activity-based" regulations. The orthogonal transcriptional switches serve as a powerful tool for dissecting the integrative role of RNAP in microbial physiology, offering meaningful implications for both fundamental studies of gene regulation and synthetic biology applications.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures








References
-
- Shabestary K, Klamt S, Link H et al. Design of microbial catalysts for two-stage processes. Nat Rev Bioeng. 2024; 2:1039–55. 10.1038/s44222-024-00225-x. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

