UBE2T promotes epithelial-mesenchymal transition and motility in oral cancer cells via induction of IL-6 expression
- PMID: 40842469
- PMCID: PMC12365892
- DOI: 10.3892/ol.2025.15219
UBE2T promotes epithelial-mesenchymal transition and motility in oral cancer cells via induction of IL-6 expression
Abstract
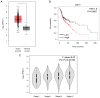
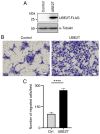
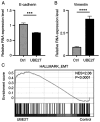
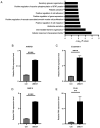
Oral squamous cell carcinoma (OSCC) is a prevalent aggressive malignancy with a high mortality rate. However, the mechanisms underlying the progression of OSCC remain to be elucidated. In the present study, bioinformatics analysis identified ubiquitin-conjugating enzyme E2 T (UBE2T) as a poor prognostic factor in head and neck cancer, showing the strongest association with cancer stage progression. Functional studies revealed that UBE2T can enhance motility and induce epithelial-mesenchymal transition (EMT) in OSCC cells. RNA sequencing and subsequent analyses demonstrated that UBE2T upregulated the expression levels of various motility- and EMT-related factors, including ankyrin repeat domain 1, endothelin-1, interleukin-6 (IL-6), matrix metalloproteinase-9 and plasminogen activator, urokinase. Gene set enrichment analysis indicated that UBE2T activates the IL-6/Janus protein tyrosine kinase (JAK)/signal transducer and activator of transcription 3 signaling pathway. Moreover, treatment of OSCC cells with IL-6 or a JAK inhibitor resulted in the induction of EMT and mesenchymal-epithelial transition, respectively, accompanied by enhanced and suppressed cancer cell motility. These results indicated that IL-6, which is upregulated by UBE2T, may be crucial for maintaining mesenchymal traits and motility in OSCC cells. Collectively, these findings suggested that the UBE2T/IL-6/JAK axis may serve as a potential therapeutic target for OSCC.
Keywords: EMT; IL-6; OSCC; UBE2T.
Copyright: © 2025 Watanabe et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
