Histopathology-based protein multiplex generation using deep learning
- PMID: 40842484
- PMCID: PMC12364712
- DOI: 10.1038/s42256-025-01074-y
Histopathology-based protein multiplex generation using deep learning
Abstract
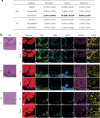
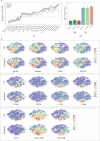
Multiplexed protein imaging offers valuable insights into interactions between tumours and their surrounding tumour microenvironment, but its widespread use is limited by cost, time and tissue availability. Here we present HistoPlexer, a deep learning framework that generates spatially resolved protein multiplexes directly from standard haematoxylin and eosin (H&E) histopathology images. HistoPlexer jointly predicts multiple tumour and immune markers using a conditional generative adversarial architecture with custom loss functions designed to ensure pixel- and embedding-level similarity while mitigating slice-to-slice variations. A comprehensive evaluation of metastatic melanoma samples demonstrates that HistoPlexer-generated protein maps closely resemble real maps, as validated by expert assessment. They preserve crucial biological relationships by capturing spatial co-localization patterns among proteins. The spatial distribution of immune infiltration from HistoPlexer-generated protein multiplex enables stratification of tumours into immune subtypes. In an independent cohort, integration of HistoPlexer-derived features into predictive models enhances performance in survival prediction and immune subtype classification compared to models using H&E features alone. To assess broader applicability, we benchmarked HistoPlexer on publicly available pixel-aligned datasets from different cancer types. In all settings, HistoPlexer consistently outperformed baseline methods, demonstrating robustness across diverse tissue types and imaging conditions. By enabling whole-slide protein multiplex generation from routine H&E images, HistoPlexer offers a cost- and time-efficient approach to tumour microenvironment characterization with strong potential to advance precision oncology.
Keywords: Computational models; Machine learning; Melanoma.
© The Author(s) 2025.
Conflict of interest statement
Competing interestsV.H.K. reports being an invited speaker for Sharing Progress in Cancer Care (SPCC) and Indica Labs; advisory board of Takeda; and sponsored research agreements with Roche and IAG, all unrelated to the current study. V.H.K. is a participant of a patent application on the assessment of cancer immunotherapy biomarkers by digital pathology, a patent application on multimodal deep learning for the prediction of recurrence risk in cancer patients and a patent application on predicting the efficacy of cancer treatment using deep learning, all of which are not directly related to the current work. G.R. and J.F.-P. are participants of a patent application on matching cells from different measurement modalities, which is not directly related to the current work. Moreover, G.R. is cofounder of Computomics GmbH, Germany, and one of its shareholders. B.B. has co-founded Navignostics, a spin-off company of the University of Zurich developing precision oncology diagnostics, and is one of its shareholders and a board member. The other authors declare no competing interests.
Figures








Update of
-
Histopathology-based Protein Multiplex Generation using Deep Learning.medRxiv [Preprint]. 2025 May 28:2024.01.26.24301803. doi: 10.1101/2024.01.26.24301803. medRxiv. 2025. Update in: Nat Mach Intell. 2025;7(8):1292-1307. doi: 10.1038/s42256-025-01074-y. PMID: 39677425 Free PMC article. Updated. Preprint.
References
-
- Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). - PubMed
-
- Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov.12, 31–46 (2022). - PubMed
-
- Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature578, 615–620 (2020). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
