Whole-transcriptome insights into follicle selection: deciphering key regulatory networks in Luxi gamecock
- PMID: 40842627
- PMCID: PMC12364954
- DOI: 10.3389/fgene.2025.1620058
Whole-transcriptome insights into follicle selection: deciphering key regulatory networks in Luxi gamecock
Abstract
Background: Luxi gamecock is a native Chinese breed recognized for its substantial body size, well-developed musculature, and aggressive behavior. Despite these advantageous traits, the breed's egg production rate remains relatively low, insufficient to meet market demands. Follicle selection plays a crucial role in determining the egg-laying performance of hens, yet research on follicle selection in Luxi gamecock is limited. In this study, RNA sequencing was performed on small yellow follicles (SYFs) and large yellow follicles (LYFs) from Luxi gamecock to identify RNA transcript expression, and subsequent RNA networks were constructed.
Methods: SYFs and LYFs were collected from 15 Luxi gamecocks and divided randomly into three biological groups. RNA was isolated to profile the expression of mRNA, lncRNA, circRNA, and miRNA. The results were validated using qRT-PCR. Functional analysis, including GO and KEGG, was conducted. Competitive endogenous RNA (ceRNA) networks were also constructed.
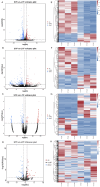
Results: A total of 1,113 mRNAs, 245 lncRNAs, 264 circRNAs, and 90 miRNAs were differentially expressed between SYFs and LYFs. qRT-PCR validation showed high consistency with the RNA-seq results. Functional enrichment indicated that these differentially expressed RNAs are associated with critical biological processes and involved in several key signaling pathways. To investigate the potential interactions among circRNAs, lncRNAs, and miRNAs, ceRNA networks were constructed.
Conclusion: This study provides a detailed characterization of the transcriptomes in SYFs and LYFs of Luxi gamecock through RNA sequencing. The functional analysis revealed that many RNAs may contribute to follicle selection. Furthermore, ceRNA networks were built to better understand the molecular mechanisms behind follicle selection. These findings shed light on the potential regulatory roles of various RNA molecules in the follicle selection of Luxi gamecock, and also uncover the interactions among them, laying a foundation for improving the breed's egg-laying performance.
Keywords: Luxi gamecock; RNA-seq; ceRNA network; differential expression RNAs; follicle selection.
Copyright © 2025 Wang.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources

