Increased reflux secondary bile acids are associated with changes to the microbiome and transcriptome in Barrett's esophagus
- PMID: 40844321
- PMCID: PMC12377100
- DOI: 10.1080/19490976.2025.2545420
Increased reflux secondary bile acids are associated with changes to the microbiome and transcriptome in Barrett's esophagus
Abstract
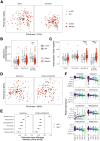
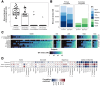
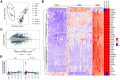
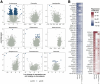
Bile acids are a major component of gastro-esophageal refluxate, thought to contribute to the development of Barrett's esophagus (BE) and esophageal adenocarcinoma (EAC). As the microbiome shifts with EAC progression and bile acids influence bacterial composition, we examined these connections in a multi-center, cross-sectional study. We analyzed biospecimens from patients undergoing endoscopy using LC-MS to quantify bile acids in gastric aspirates, 16S rRNA sequencing for tissue microbiome profiling, and RNA sequencing on BE or cardia tissue. Among 153 patients (52 controls, 101 BE: 50 no dysplasia, 10 indefinite, 17 low-grade dysplasia, 17 high-grade dysplasia, and 7 EAC), we observed increased Streptococcus in BE tissue; dysplasia and EAC were associated with more Lactobacillus and decreased Actinomyces and other genera. Refluxate bile acids were mainly conjugated, indicating minimal bacterial metabolism, while BE patients had elevated secondary bile acid levels. Streptococcus correlated with upregulation of IL6, FGF2, and HGF, and decreased Actinomyces showed the most associations with gene expression, including the oxidative phosphorylation pathway. We identified two distinct BE gene expression clusters independent of histology, bile acid, or microbiome composition. These findings suggest bile acids shape the BE microbiome and associate with gene expression changes potentially relevant to EAC development.
Keywords: Barrett’s esophagus; bile acids; esophageal adenocarcinoma; microbiome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Ward JBJ, Lajczak NK, Kelly OB, O’Dwyer AM, Giddam AK, Ní Gabhann J, Franco P, Tambuwala MM, Jefferies CA, Keely S, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol Gastrointest Liver Physiol. 2017;312(6):G550–G558. doi: 10.1152/ajpgi.00256.2016. - DOI - PubMed
-
- Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee M, Lee Y, Friedman R, Asfaha S, Dubeykovskaya Z, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21(1):36–51. doi: 10.1016/j.ccr.2011.12.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
