Characterization of the glycoproteins of fish and amphibian influenza B-like viruses
- PMID: 40845102
- PMCID: PMC12372866
- DOI: 10.1126/sciadv.ady8610
Characterization of the glycoproteins of fish and amphibian influenza B-like viruses
Abstract
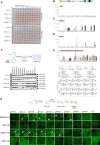
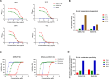
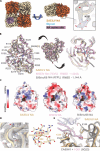
Influenza-like virus sequences previously identified in fish and amphibians cluster as a sister clade of influenza B viruses but remain largely uncharacterized. We demonstrate that salamander influenza-like virus (SILV) hemagglutinin (HA) is functionally divergent from influenza B virus HA and does not bind to α2,3- and α2,6-linked sialic acids. However, the HAs of Siamese algae-eater influenza-like virus (SAEILV) and chum salmon influenza-like virus (CSILV) bind to α2,3-linked sialic acid. Furthermore, SAEILV HA binds to sialyated Lewis X, is activated by human airway enzymes, and is fusogenic over a broad pH range. SAEILV neuraminidase (NA) has a highly conserved active site and a similar structure to other known NAs. We also determined the cryo-electron microscopy structure of the HA of a previously described virus from the same sister clade, the Wuhan spiny eel influenza virus (WSEIV). No cross-reactive antibodies against these HAs or NAs were found in human serum, suggesting that humans are immunologically naïve to these viruses.
Figures



Update of
-
Characterization of the glycoproteins of novel fish influenza B-like viruses.bioRxiv [Preprint]. 2025 May 8:2025.05.08.652883. doi: 10.1101/2025.05.08.652883. bioRxiv. 2025. Update in: Sci Adv. 2025 Aug 22;11(34):eady8610. doi: 10.1126/sciadv.ady8610. PMID: 40654808 Free PMC article. Updated. Preprint.
Similar articles
-
Characterization of the glycoproteins of novel fish influenza B-like viruses.bioRxiv [Preprint]. 2025 May 8:2025.05.08.652883. doi: 10.1101/2025.05.08.652883. bioRxiv. 2025. Update in: Sci Adv. 2025 Aug 22;11(34):eady8610. doi: 10.1126/sciadv.ady8610. PMID: 40654808 Free PMC article. Updated. Preprint.
-
Functionality of the putative surface glycoproteins of the Wuhan spiny eel influenza virus.Nat Commun. 2021 Oct 25;12(1):6161. doi: 10.1038/s41467-021-26409-2. Nat Commun. 2021. PMID: 34697321 Free PMC article.
-
Binding antibody titers against the hemagglutinin and neuraminidase correlate with protection against medically attended influenza A and B disease.J Virol. 2025 Jun 17;99(6):e0039125. doi: 10.1128/jvi.00391-25. Epub 2025 May 13. J Virol. 2025. PMID: 40358209 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
-
Neuraminidase inhibitors for preventing and treating influenza in adults and children.Cochrane Database Syst Rev. 2014 Apr 10;2014(4):CD008965. doi: 10.1002/14651858.CD008965.pub4. Cochrane Database Syst Rev. 2014. PMID: 24718923 Free PMC article.
References
-
- D. M. Knipe, P. Howley, Fields Virology 6th Edition (Lippincott Williams & Wilkins, 2013).
-
- Halwe N. J., Hamberger L., Sehl-Ewert J., Mache C., Schön J., Ulrich L., Calvelage S., Tönnies M., Fuchs J., Bandawane P., Loganathan M., Abbad A., Carreño J. M., Bermúdez-González M. C., Simon V., Kandeil A., El-Shesheny R., Ali M. A., Kayali G., Budt M., Hippenstiel S., Hocke A. C., Krammer F., Wolff T., Schwemmle M., Ciminski K., Hoffmann D., Beer M., Bat-borne H9N2 influenza virus evades MxA restriction and exhibits efficient replication and transmission in ferrets. Nat. Commun. 15, 3450 (2024). - PMC - PubMed
-
- Osterhaus A. D., Rimmelzwaan G. F., Martina B. E., Bestebroer T. M., Fouchier R. A., Influenza B virus in seals. Science 288, 1051–1053 (2000). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

