Early lineage segregation of primary myotubes from secondary myotubes and adult muscle stem cells
- PMID: 40846698
- PMCID: PMC12374003
- DOI: 10.1038/s41467-025-61767-1
Early lineage segregation of primary myotubes from secondary myotubes and adult muscle stem cells
Abstract
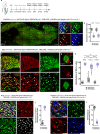
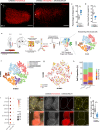
Myogenesis in amniotes occurs in two waves. Primary myotubes express slow myosin (often with fast myosin) and likely act as scaffolds for secondary myotubes, which express only fast myosin. The embryonic origins and relationships of these lineages, and their connection to satellite cells, remain unknown. Here, we combine a TCF-LEF/β-catenin signaling reporter with precise in vivo electroporation in avian embryos to trace limb muscle progenitors from early migration to fetal stages. We identify two distinct progenitor populations that coexist from the onset: reporter-positive cells give rise exclusively to primary myotubes, while reporter-negative cells generate secondary myotubes and satellite cells. We also reveal a previously unrecognized role for TCF-LEF/β-catenin signaling in spatially organizing the primary lineage via Cxcr4-mediated control of myoblast migration. These findings redefine the developmental origin of myogenic lineages, resolve a longstanding question in muscle biology, and provide a molecular framework for investigating how muscle fiber diversity emerges and how distinct lineages contribute to the functional specialization of skeletal muscle.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests
Figures



References
-
- Christ, B. & Ordahl, C. Early stages of chick somite development. Anat. Embryol.191, 381–396 (1995). - PubMed
-
- Bryson-Richardson, R. J. & Currie, P. D. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008 989, 632–646 (2008). - PubMed
-
- Hirst, C. E. C. E. & Marcelle, C. The avian embryo as a model system for skeletal myogenesis. Results and Problems in Cell Differentiation56, 99–122 (Springer, 2015). - PubMed
-
- Scaal, M. & Marcelle, C. Chick muscle development. Int. J. Dev. Biol.62, 127–136 (2018). - PubMed
-
- Gros, J., Scaal, M. & Marcelle, C. A two-Step mechanism for myotome formation in chick. Dev. Cell6, 875-882 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

