SRAS1.1 E3 ligase mediates DSK2A degradation to regulate autophagy and drought tolerance in Arabidopsis
- PMID: 40847257
- PMCID: PMC12508185
- DOI: 10.1038/s44319-025-00556-9
SRAS1.1 E3 ligase mediates DSK2A degradation to regulate autophagy and drought tolerance in Arabidopsis
Abstract
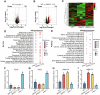
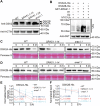
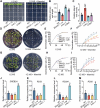
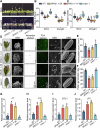
Drought stress significantly impacts plant growth and productivity, requiring complex adaptive responses to ensure survival. In eukaryotes, autophagy and the ubiquitin-proteasome system (UPS) are critical pathways for maintaining cellular homeostasis under stress. While their interaction is well-studied in animals, it remains poorly understood in plants, particularly under drought conditions. Here, we identify the E3 ubiquitin ligase SRAS1.1 as a key regulator of selective autophagy and drought tolerance in Arabidopsis, mediating its function through the ubiquitination and degradation of the autophagy receptor DSK2A. Loss of SRAS1.1 enhances drought tolerance by reducing water loss, increasing survival rates, and accelerating flowering. SRAS1.1 directly interacts with and ubiquitinates the autophagy receptor DSK2A, promoting its degradation via the 26S proteasome. Notably, under drought stress, SRAS1.1 relocates from the nucleus to the cytoplasm, associates with autophagosomes, and modulates autophagy-related gene expression and BES1 accumulation. These findings provide novel insights into UPS-autophagy crosstalk in plants and highlight SRAS1.1 as a promising target for genetic engineering to develop drought-resilient crops and to advance sustainable agriculture.
Keywords: Autophagy; DSK2A; Drought Stress; E3 Ubiquitin Ligase; Stress Tolerance.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures






References
-
- Chen X, Wang T, Rehman AU, Wang Y, Qi J, Li Z, Song C, Wang B, Yang S, Gong Z (2021b) Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J Integr Plant Biol 63:494–509 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

