Landscape analysis of m5C modification regulators unveils DNMT1-mediated dysregulated pyrimidine metabolism in hepatocellular carcinoma
- PMID: 40847321
- PMCID: PMC12372286
- DOI: 10.1186/s13148-025-01956-3
Landscape analysis of m5C modification regulators unveils DNMT1-mediated dysregulated pyrimidine metabolism in hepatocellular carcinoma
Abstract
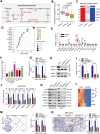
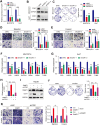
The 5-methylcytosine (m5C) post-transcriptional modification has been linked with the development and progression of a variety of cancers. However, its specific functions and their underlying mechanisms are poorly understood in hepatocellular carcinoma (HCC). The present study showed abnormally increased levels of m5C modifications in HCC that were positively correlated with both HCC progression and worse patient prognosis. Landscape profiling of metabolic characteristics showed dysregulation of pyrimidine metabolism mediated by DNA methyltransferases 1 (DNMT1), and cyclin-dependent kinase 1 (CDK1) was identified as a downstream effector upregulated by DNMT1 in an m5C-dependent manner, with CDK1 promoting pyrimidine metabolism. Knockdown of DNMT1 or CDK1 was found to reduce the proliferation, invasion, and migration of HCC cells in vitro. Moreover, pharmacological targeting of the DNMT1/CDK1/pyrimidine metabolism axis with specific inhibitors effectively suppressed tumor progression in HCC model mice. These findings demonstrated the landscape profiles of m5C-related metabolic features in HCC, showing stabilization of CDK1 mRNA by DNMT1-mediated m5C modification, resulting in the promotion of pyrimidine metabolism, a crucial feature of HCC progression. These insights highlight the therapeutic potential of targeting the DNMT1/CDK1/pyrimidine metabolism axis as a strategy for combating HCC.
Keywords: Cyclin-dependent protein kinase 1; DNA-methyltransferase 1; HCC; Pyrimidine metabolism; m5C modification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests. Ethics approval and consent to participate: The collection of specimens of clinical patients in this study was approved by the Research Ethics Committee of Zhongshan Hospital. Consent for publication: Not applicable.
Figures





References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7: 6. - PubMed
-
- Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–62. - PubMed
-
- Berggren KA, Schwartz RE, Kleiner RE, Ploss A. The impact of epitranscriptomic modifications on liver disease. Trends Endocrinol Metab. 2024;35:331–46. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

