Molecular landscape, subtypes, and therapeutic vulnerabilities of central nervous system solitary fibrous tumors
- PMID: 40849425
- PMCID: PMC12375123
- DOI: 10.1038/s41467-025-63039-4
Molecular landscape, subtypes, and therapeutic vulnerabilities of central nervous system solitary fibrous tumors
Abstract
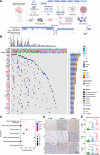
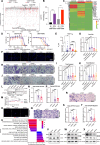
Solitary fibrous tumors (SFTs) of the central nervous system (CNS) are rare, aggressive mesenchymal neoplasms with high recurrence and metastasis rates. Here, we perform a comprehensive multi-omics analysis of 189 cases of CNS SFTs integrating 94 whole genome sequencing, 88 transcriptomics, 7 single-nucleus RNA sequencing and 3 spatial transcriptome sequencing. We find that receptor tyrosine kinase mutations are significantly more prevalent besides the widespread NAB2-STAT6 fusion and correlate with tumor grade. Transcriptomic analysis reveals four molecular subtypes-classical, neural-like, inflamed and migratory-each associated with distinct clinical and biological characteristics. Single-nucleus RNA sequencing identifies five tumor cell states, with the SFT_classical state serving as a precursor to other states influenced by hypoxia and inflammation. Notably, we identify FER kinase as a key therapeutic target, with FER inhibition significantly reducing tumor cell proliferation, migration and invasion. These findings provide important insights into CNS SFT biology and suggest potential therapeutic strategies for this challenging tumor type.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Carneiro, S. S., Scheithauer, B. W., Nascimento, A. G., Hirose, T. & Davis, D. H. Solitary fibrous tumor of the meninges: a lesion distinct from fibrous meningioma: a clinicopathologic and immunohistochemical study. Am. J. Clin. Pathol.106, 217–224 (1996). - PubMed
-
- Sughrue, M. E. et al. The relevance of simpson grade I and II resection in modern neurosurgical treatment of World Health Organization grade I meningiomas: clinical article. J. Neurosurg.113, 1029–1035 (2010). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

