H4K79 and H4K91 histone lactylation, newly identified lactylation sites enriched in breast cancer
- PMID: 40849487
- PMCID: PMC12374308
- DOI: 10.1186/s13046-025-03512-6
H4K79 and H4K91 histone lactylation, newly identified lactylation sites enriched in breast cancer
Abstract
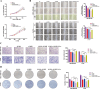
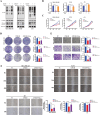
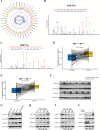
Metabolic reprogramming and epigenetic modification are two hallmarks of cancer. Protein lysine lactylation (Kla) is a novel type of glycolysis lactate-triggered posttranslational modification. However, the role of Kla in breast cancer (BC) remains largely unknown. Here, western blot, and immunohistochemical (IHC) staining of BC tissues revealed that global Kla levels were upregulated in BC tissues, and high levels of Kla were correlated with poor prognosis of patients with BC. A series of in vitro and in vivo assays demonstrated that interruption of glycolysis by lactate dehydrogenase (LDH) inhibitor or silencing LDHA and LDHB repressed the malignant behaviors of BC cells. Moreover, 4D label-free quantitative lactylproteomics analysis of BC tissues and cells revealed that lactylated proteins widely existed in several subcellular compartments and were closely associated with various cancer-related biological processes. Notably, two previously unresearched sites of histone lactylation, H4K79 lactylation (H4K79la) and H4K91 lactylation (H4K91la), were identified to be hyperlactylated in cancer tissues and cells. Glycolytic genes, such as lactate dehydrogenase A (LDHA), phosphoglycerate kinase 1 (PGK1), and hexokinase 1 (HK1) were identified to be the potential candidate genes epigenetically regulated by H4K79la and H4K91la by intersecting through chromatin immunoprecipitation sequencing (ChIP-seq), RNA sequencing (RNA-seq), and TCGA-BRCA database. Pharmacological inhibition of glycolysis downregulated H4K79 and H4K91 lactylation and suppressed the expression of glycolytic genes, whereas treatment with sodium lactate exhibited the opposite effects. Additionally, E1A-binding protein p300 (P300) acted as lysine lactyltransferase to regulate H4K79la and H4K91la, and control the transcription and expression of downstream glycolytic genes in BC cells. The results revealed an intriguing positive feedback loop formed by glycolysis/H4K79la/H4K91la/glycolytic genes in BC, highlighting the relationship between metabolic reprogramming and epigenetic regulation. These findings provide new therapeutic targets for patients with BC.
Keywords: Breast cancer; Epigenetic modification; Glycolysis; Lactylation; Post-translational modifications.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: The Medical Ethics Committee of Harbin Medical University Cancer Hospital approved this study (KY2023-84). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures








References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

