Characteristics of gut and lung microbiota in patients with lung masses and their relationship with clinical features
- PMID: 40849619
- PMCID: PMC12374282
- DOI: 10.1186/s12866-025-04325-5
Characteristics of gut and lung microbiota in patients with lung masses and their relationship with clinical features
Abstract
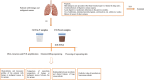
Objectives: The exploration of how dysbiosis relates to lung masses is still nascent, with few studies focusing on the microbial characteristics across various sites. Therefore, we categorized the microbiota into feces and bronchoalveolar fluid (BALF) groups to compare microbial characteristics between benign and malignant masses, analyze their clinical correlations, and develop predictive models for lung cancer.
Methods: A total of 238 fecal samples and 34 BALF samples were collected from patients with benign and malignant masses and then analyzed by 16 SrRNA. We explored the distinct composition of the gut and lung microbiota and their associations with clinical features. The diagnostic models were constructed using microbial features identified through two approaches: random forest algorithm with five-fold cross-validation and comparative analysis of significantly differential taxa. The performance evaluation was subsequently conducted using receiver operating characteristic (ROC) analysis.
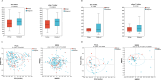
Results: There was no significant difference in α-and β-diversity between feces and BALF groups. The relative abundance of Lachnospiraceae_NK4A136_group (P = 0.003232) and Erysipelotrichaceae_UCG-003 (P = 0.01316) in feces group and Proteobacteria (P = 0.03654) in BALF group were significantly increased in lung cancer patients. We also found Bacteroides (P = 0.01458) was abundant in NSCLC than those of SCLC in feces group, while the BALF group was dominated by norank_c_Cyanobacteria (P = 0.03384). Smoking history appeared to be related to the distribution of microbiota, with enrichment of Parabacteroides (P = 0.02054) in feces and Prevotella_1 (P = 0.03286) in BALF. Furthermore, the patients with Sellimonas (P = 0.04148) in feces and Alloprevotella (P = 0.04283) in BALF seemed to have better response to chemotherapy combined with immunotherapy. For differentiating benign and malignant masses, the combination of Megasphaera and norank_p__Saccharibacteria in BALF demonstrated superior predictive performance, with an AUC reaching 0.8 (95% CI 0.59-1).
Conclusion: The microbiota composition significantly differed between benign and malignant masses in both fecal and BALF groups, with minimal evidence supporting microbial migration between these two sites. Our findings suggest that BALF microbiota may serve as a more reliable biomarker for lung masses classification, offering valuable insights for early diagnosis and clinical decision-making.
Keywords: Clinical features; Gut microbiota; Lung masses; Lung microbiota.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All participants were informed of the study and written consent was obtained prior to enrollment in the study. This study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University (approval number: B2020-019R). This study was conducted in compliance with the principles of the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
[Relationship Between Different Traditional Chinese Medicine Syndrome Types and Gut Microbiota in Patients With Type 2 Diabetes Mellitus].Sichuan Da Xue Xue Bao Yi Xue Ban. 2025 Mar 20;56(2):389-399. doi: 10.12182/20250360507. Sichuan Da Xue Xue Bao Yi Xue Ban. 2025. PMID: 40599291 Free PMC article. Chinese.
-
Microbiome and metabolome patterns after lung transplantation reflect underlying disease and chronic lung allograft dysfunction.Microbiome. 2024 Oct 9;12(1):196. doi: 10.1186/s40168-024-01893-y. Microbiome. 2024. PMID: 39385282 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
MeSH terms
Grants and funding
- ZD2021CY001/Shanghai Municipal Science and Technology Major Project
- 20Z11901000, 20DZ2261200, 20XD1401200, 22Y11900800/Science and Technology Commission of Shanghai Municipality
- shslczdzk02201/Shanghai Municipal Key Clinical Specialty
- ZY(2021-2023)-0207-01/Shanghai Municipal Health Commission and Shanghai Municipal Administrator of Traditional Chinese Medicine
- 81401877, 82130001,82272243/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical

