Elevated levels of Letm1 drives mitochondrial dysfunction and cardiomyocyte stress-mediated apoptosis in cultured cardiomyocytes
- PMID: 40849623
- PMCID: PMC12374280
- DOI: 10.1186/s12964-025-02378-7
Elevated levels of Letm1 drives mitochondrial dysfunction and cardiomyocyte stress-mediated apoptosis in cultured cardiomyocytes
Abstract
Background: Cardiac ischemia, a predominant cause of heart failure, is marked by profound mitochondrial dysfunction, dysregulated ion homeostasis, and maladaptive cellular remodeling, all of which compromise cardiac performance. The mitochondrial inner membrane protein Leucine zipper-EF-hand containing Transmembrane Protein 1 (Letm1), implicated in Wolf-Hirschhorn Syndrome, is essential for mitochondrial function. Although genetic alterations in Letm1 are linked to cardiomyopathies, its specific contributions to cardiac pathophysiology, particularly in the context of ischemic heart disease, remain poorly defined. This study aims to elucidate the role of Letm1 in ischemic cardiac pathology and its mechanistic impact on cardiomyocyte function.
Methods: Letm1 expression was assessed in human and murine models of heart failure due to ischemic cardiomyopathy (ICM) and cardiac hypertrophy. Letm1 was overexpressed in neonatal rat ventricular cardiomyocytes, adult mouse cardiomyocytes, and human induced pluripotent stem cell (iPSC)-derived cardiomyocytes to study mitochondrial function (Seahorse assays), structural and molecular remodeling (fluorescence microscopy, transmission electron microscopy (TEM), qPCR, immunoblotting), transcriptomic/proteomic profiles, calcium handling and electrophysiology (patch-clamp), autophagic flux (Bafilomycin A1, LC3-RFP-GFP), and cell survival.
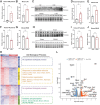
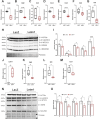
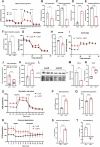
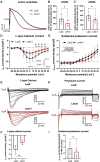
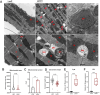
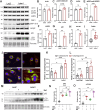
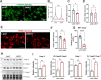
Results: Letm1 was markedly upregulated in ICM in both human and murine hearts, but unchanged in hypertrophic heart failure. Overexpression of Letm1 in cardiomyocytes resulted in profound mitochondrial dysfunction, including downregulation of oxidative phosphorylation (OXPHOS) genes, impaired membrane potential, reduced ATP output, increased proton leak, and elevated ROS levels. A metabolic shift toward glycolysis was observed, accompanied by reduced fatty acid oxidation. Electron microscopy revealed mitochondrial fragmentation, mitophagic vesicles, and sarcomeric disarray. Transcriptomic and proteomic analyses highlighted dysregulation of genes linked to mitochondrial organization, ion transport, and autophagy. Electrophysiologically, Letm1 reduced L-type Ca2+ current density and significantly shortened action potential duration, leading to impaired contractility. Letm1 overexpression activated upstream autophagy regulators (AMPK, ULK1) and enhanced LC3-II and p62 accumulation, but autophagic flux was impaired, as confirmed by LC3-RFP-GFP reporter and exacerbated by Bafilomycin A1 treatment. This dysregulated autophagy was coupled with mitochondrial stress, increased apoptosis (cleaved caspases), and reduced cardiomyocyte viability.
Conclusion: This study indicates that Letm1 upregulation drives mitochondrial dysfunction, electrophysiology alterations, and activation of autophagy and apoptosis, culminating in cardiomyocyte injury in ischemic cardiomyopathy. By disrupting OXPHOS, calcium handling, and cell survival pathways, Letm1 contributes to ischemic remodeling and cardiac dysfunction. Targeting Letm1 presents a promising therapeutic strategy to alleviate ischemic damage and preserve cardiac function.
Keywords: Arrhythmias; Cardiomyocytes; Hypertrophy; Letm1; Mitochondrial metabolism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal experiments were performed according to international, institutional and governmental ethical guidelines. LAD mouse experiment was approved by the Ministry of Energy Transition, Agriculture, Environment, Nature and Digitalization (MELUND) of the state of Schleswig-Holstein (96 − 11/20) and ORAB mouse experiment was approved by the authorities of the Regierungspräsidium Karlsruhe (G-174/23). The use of human tissue samples conforms to the declaration of Helsinki and was approved by the ethical committee of the medical school of the Georg-August-University, Göttingen. Written informed consent was received from all participants prior to inclusion. Consent for publication: All authors declare consent for publication. Competing interests: The authors declare no competing interests.
Figures

References
-
- Vaduganathan M, Mensah GA, Turco JV, Fuster V, Roth GA. The global burden of cardiovascular diseases and risk: A compass for future health. J Am Coll Cardiol. 2022;80:2361–71. 10.1016/j.jacc.2022.11.005. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

