Reverse vaccinology-based identification and in silico characterization of immunogenic membrane proteins of Salmonella Typhimurium as novel vaccine targets against multidrug-resistant infections
- PMID: 40849649
- PMCID: PMC12375273
- DOI: 10.1186/s12866-025-04124-y
Reverse vaccinology-based identification and in silico characterization of immunogenic membrane proteins of Salmonella Typhimurium as novel vaccine targets against multidrug-resistant infections
Abstract
Background: Salmonella enterica serovar Typhimurium (S. Typhimurium) is a leading cause of salmonellosis, gastroenteritis, sepsis, and reactive arthritis. Transmission primarily occurs through contaminated water, eggs, meat, and dairy products. The disease disproportionately affects developing nations, where young children, the elderly, and immunocompromised individuals face high risks of severe morbidity and mortality. Its ability to evade host immune defenses and acquire multidrug resistance (MDR) exacerbates global public health challenges. Currently, no licensed human vaccine is available, underscoring the urgent need for targeted vaccine development.
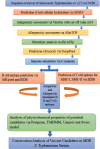
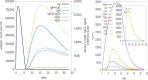
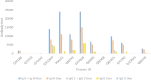
Methods: This study utilized a reverse vaccinology approach and in silico strategies to identify highly immunogenic membrane proteins as potential vaccine candidates. The complete proteome of S. Typhimurium was screened for membrane-associated candidates using the SOSUI server. Antigenicity was evaluated using VaxiJen v2.0 (threshold ≥ 0.9), and allergenicity was assessed using AllerTOP v1.1. To ensure vaccine safety, homologous proteins were excluded based on PSI-BLAST analysis against the human proteome, and toxicity was predicted using ToxinPred. The immunogenic potential was further evaluated through C-ImmSim immune simulation software. B-cell and T-cell epitopes were predicted using ABCpred and the Immune Epitope Database (IEDB). Physicochemical characteristics were analyzed with ProtParam and TMHMM 2.0. Finally, BLASTp analysis was used to confirm the conservation of the selected proteins across MDR clinical isolates.
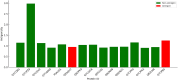
Results: Nine membrane proteins were prioritized based on strong antigenicity, non-allergenicity, non-toxicity, favorable epitope profiles, and physicochemical stability. All proteins were highly conserved in MDR isolates, supporting their utility for broad-spectrum vaccine development.
Conclusion: These targets show promising potential for developing a broadly protective multi-epitope vaccine against S. Typhimurium. However, in vitro and in vivo experimental validation is essential to confirm their immunogenicity and protective efficacy.
Keywords: In silico analysis; Salmonella Typhimurium; B-cell epitopes; Immunogenicity; Multidrug-resistance; Reverse vaccinology; T-cell epitopes; Vaccine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Screening of novel therapeutic targets and chimeric vaccine construction against antibiotic-resistant Yersinia Enterocolitica.Front Immunol. 2025 Jul 4;16:1555248. doi: 10.3389/fimmu.2025.1555248. eCollection 2025. Front Immunol. 2025. PMID: 40688091 Free PMC article.
-
Development of a multi-epitope vaccine candidate targeting blood-stage of malaria through immunoinformatics approach.Hum Immunol. 2025 Jul;86(4):111346. doi: 10.1016/j.humimm.2025.111346. Epub 2025 Jul 15. Hum Immunol. 2025. PMID: 40669099
-
Immunoinformatics-Based development of a Multi-Epitope vaccine candidate targeting coinfection by Klebsiella pneumoniae and Acinetobacter baumannii.BMC Infect Dis. 2025 Jul 3;25(1):894. doi: 10.1186/s12879-025-11242-5. BMC Infect Dis. 2025. PMID: 40610881 Free PMC article.
-
Combatting Salmonella: a focus on antimicrobial resistance and the need for effective vaccination.BMC Infect Dis. 2025 Jan 20;25(1):84. doi: 10.1186/s12879-025-10478-5. BMC Infect Dis. 2025. PMID: 39833704 Free PMC article.
-
Immunoinformatics-Driven Design of a Multi-Epitope Vaccine Targeting Simian Virus VP1 Major Capsid Protein for Oncogenic Viral Infection Prevention.Rev Med Virol. 2025 Sep;35(5):e70065. doi: 10.1002/rmv.70065. Rev Med Virol. 2025. PMID: 40781525 Review.
References
-
- Jafari Najaf Abadi MH, Abdi Abyaneh F, Zare N, Zamani J, Abdoli A, Aslanbeigi F, et al. Silico design and immunoinformatics analysis of a chimeric vaccine construct based on Salmonella pathogenesis factors. Microb Pathog. 2023;180:106130. 10.1016/j.micpath.2023.106130. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

