KDM4 Orchestrates Epigenomic Remodeling of Senescent Cells and Potentiates the Senescence-Associated Secretory Phenotype
- PMID: 40849896
- PMCID: PMC12507408
- DOI: 10.1111/acel.70194
KDM4 Orchestrates Epigenomic Remodeling of Senescent Cells and Potentiates the Senescence-Associated Secretory Phenotype
Abstract
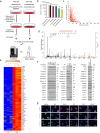
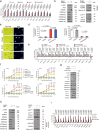
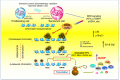
Cellular senescence restrains the expansion of neoplastic cells through several layers of regulation. We report that the histone H3-specific demethylase KDM4 is expressed as human stromal cells undergo senescence. In clinical oncology, upregulated KDM4 and diminished H3K9/H3K36 methylation correlate with poorer survival of patients with prostate cancer after chemotherapy. Global chromatin accessibility mapping via assay for transposase-accessible chromatin with high-throughput sequencing, and expression profiling through RNA sequencing, reveals global changes of chromatin openness and spatiotemporal reprogramming of the transcriptomic landscape, which underlie the senescence-associated secretory phenotype (SASP). Selective targeting of KDM4 dampens the SASP of senescent stromal cells, promotes cancer cell apoptosis in the treatment-damaged tumor microenvironment, and prolongs survival of experimental animals. Our study supports dynamic changes of H3K9/H3K36 methylation during senescence, identifies an unusually permissive chromatin state, and unmasks KDM4 as a key SASP modulator. KDM4 targeting presents a new therapeutic avenue to manipulate cellular senescence and limit its contribution to age-related pathologies, including cancer.
Keywords: H3K9/H3K36 demethylation; KDM4; age‐related pathology; aging; cellular senescence; epigenetic modification.
© 2025 The Author(s). Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Acosta, J. C. , O'Loghlen A., Banito A., Guijarro M. V., Hernando E., and Gil J.. 2008. “Chemokine Signaling via the CXCR2 Receptor Reinforces Senescence.” Cell 133, no. 6: 1006–1018. - PubMed
-
- Bae, V. L. , Jackson‐Cook C. K., Maygarden S. J., Plymate S. R., Chen J., and Ware J. L.. 1998. “Metastatic Sublines of an SV40 Large T Antigen Immortalized Human Prostate Epithelial Cell Line.” Prostate 34, no. 4: 275–282. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

