Virulence factors are preserved within carbapenem-resistant Acinetobacter baumannii clades
- PMID: 40849900
- PMCID: PMC12377095
- DOI: 10.1080/21505594.2025.2542489
Virulence factors are preserved within carbapenem-resistant Acinetobacter baumannii clades
Abstract
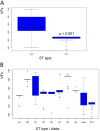
Carbapenem-resistant Acinetobacter baumannii (CRAB) is an important threat to hospitalized patients. The virulence of A. baumannii varies between strains and geographic locations, which is believed to be related to its genetic plasticity. A comprehensive evaluation of virulence factor (VF) content of CRAB across genetically related clones is lacking. Therefore, we aimed to determine the evolution of VFs among 246 CRAB isolates belonging to major STs, ST2 and ST3. We used WGS to assess 136 VFs and found that 110 VFs were present in most isolates (49 VFs were universally present, 61 were ubiquitous) and 25 occurred sporadically. The distribution of the 25 sporadic VF genes was homogenous in ST3 but heterogeneous in ST2. Within ST2, we found high intra-clade homogeneity and high inter-clade heterogeneity of VFs. The homogeneity of VF content in ST2 clades and its uniformity in ST3 suggest a distant evolutionary segregation of VFs at the root of ST/clade divergence. VF content reflects remote events resulting in minimal variation between isolates belonging to the same ST/clade. Thus, the alleged differences in virulence of CRAB strains between geographical locations reflect differences in clonal distribution.
Keywords: Acinetobacter baumannii; antibiotic resistance; carbapenem resistance; evolution; virulence factors.
Conflict of interest statement
Yehuda Carmeli received consultancy fees from MSD, Qpex, Pfizer, Roche, and Enlivex and payment for lectures from Pfizer. Emanuele Durante‐Mangoni received grants from Pfizer, Shionogi, Advanz Pharma, Infectopharm, and Angelini, and payment for lectures from Pfizer, Shionogi, Advanz Pharma, Infectopharm, Angelini, Abbvie, and Trx and participated on a data safety monitoring board or advisory board of Roche, Genentech, and Pfizer. George Daikos received honoraria for lectures from Pfizer, MSD and received support to attend meetings from Pfizer and participated on an advisory board of Vitaris, MSD, and Pfizer. Dafna Yahav received grants from Pfizer and Shionogi. All other authors declared no competing interests in this work.
Figures

References
-
- Bouvet PJM, Grimont PAD.. taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov. Ac. Int J Systematic Bacteriol. 1986;36(2):228–240. doi: 10.1099/00207713-36-2-228 - DOI
-
- World Health Organization . Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. Available from 2017. doi: 10.1002/wcc.119 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
