Oxidative stress-induced ZEB1 acetylation drives a hybrid epithelial-mesenchymal phenotype and promotes lung metastasis in triple-negative breast cancer
- PMID: 40850192
- PMCID: PMC12398892
- DOI: 10.1016/j.redox.2025.103834
Oxidative stress-induced ZEB1 acetylation drives a hybrid epithelial-mesenchymal phenotype and promotes lung metastasis in triple-negative breast cancer
Abstract
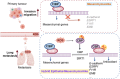
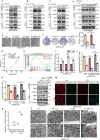
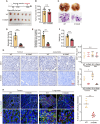
While epithelial-mesenchymal plasticity (EMP) drives cancer metastasis, its regulation by redox dynamics remains poorly understood. Herein, we identified an oxidative stress-responsive CBP/SIRT1 axis that coordinated ZEB1 acetylation at K1108 to promote lung metastasis in triple-negative breast cancer (TNBC). Mechanistically, the biochemical and functional analyses revealed that the dual-acetyltransferase CBP, through stabilization and autoacetylation by oxidative stress, formed a dynamic partnership with SIRT1 to execute precision lysine modification. This post-translational rheostat triggered the functional metamorphosis of ZEB1. During this process, ZEB1 dissociation from the transcriptional corepressor CtBP, while recruiting CBP, converts ZEB1 into a transcriptional activator of epithelial genes. The resulting hybrid epithelial‒mesenchymal phenotype orchestrated dual metastatic competence-maintaining stromal interaction capacity through partial epithelial‒mesenchymal transition (EMT) while establishing NADPH-driven redox supremacy to circumvent ferroptosis. Importantly, this acetyl switch of ZEB1 revealed a metastasis-specific therapeutic vulnerability in TNBC. Our work thus highlighted ZEB1 acetylation as a redox-interpreted mechanism coupling phenotypic plasticity with stress resistance, proposing targeted disruption of this protein post-translational modification (PTM) circuit as a precision strategy against metastatic progression.
Keywords: Acetylation; Hybrid epithelial-mesenchymal phenotype; Lung metastasis; Oxidative stress; TNBC; ZEB1.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest None declared.
Figures





References
-
- Bray F., et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024;74(3):229–263. - PubMed
-
- Lambert A.W., Weinberg R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer. 2021;21(5):325–338. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

