High-Throughput Nanorheology of Living Cells Powered by Supervised Machine Learning
- PMID: 40852088
- PMCID: PMC12370168
- DOI: 10.1002/aisy.202400867
High-Throughput Nanorheology of Living Cells Powered by Supervised Machine Learning
Abstract
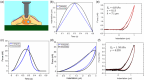
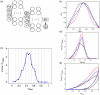
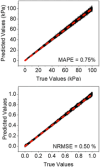
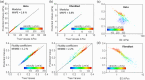
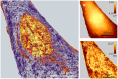
Atomic force microscopy (AFM) is extensively applied to measure the nanomechanical properties of living cells. Despite its popularity, some applications on mechanobiology are limited by the low throughput of the technique. Currently, the analysis of AFM-nanoindentation data is performed by model fitting. Model fitting is slow, data intensive, and prone to error. Herein, a supervised machine-learning regressor is developed for transforming AFM force-distance curves into nanorheological behavior. The method reduces the computational time required to process a force volume of a cell made of 2.62 × 105 curves from several hours to minutes. In fact, the regressor increases the throughput by 50-fold. The training and the validation of the regressor are performed by using theoretical curves derived from a contact mechanics model that combined power-law rheology with bottom effect corrections and functional data analysis. The regressor predicts the modulus and the fluidity coefficient of mammalian cells with a relative error below 4%.
Keywords: atomic force microscopies; mammalian cells; mechanobiologies; nanoindentations; nanorheologies.
© 2025 The Author(s). Advanced Intelligent Systems published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Efremov Y. M., Okajima T., Raman A., Soft Matter 2020, 16, 64. - PubMed
-
- Garcia R., Chem. Soc. Rev. 2020, 49, 5850.
-
- Flormann D. A. D., Anton C., Pohland M. O., Bautz Y., Kaub K., Terriac E., Schäffer T. E., Rheinlaender J., Janshoff A., Ott A., Lautenschläger F., Front. Phys. 2021, 9, 711860.
-
- Li M., Liu L., Xiao X., Xi N., Wang Y., IEEE Trans. Nanobiosci. 2016, 15, 398.
LinkOut - more resources
Full Text Sources
Miscellaneous
