Understanding the interaction between melatonin and bifidobacteria
- PMID: 40852128
- PMCID: PMC12367468
- DOI: 10.20517/mrr.2025.10
Understanding the interaction between melatonin and bifidobacteria
Abstract
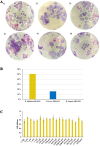
Aim: The human gastrointestinal tract is home to a complex and dynamic microbial community, known as the gut microbiota, which begins to form at birth and evolves throughout life. Among the factors influencing its initial development, breastfeeding is one of the most important. Human milk is a chemically complex body fluid, including hormones, like melatonin, which is involved in regulating the sleep-wake cycle, helping to establish the newborn's circadian rhythm. In the current study, the molecular interactions between human melatonin and a bifidobacteria-rich infant gut microbiota were explored. Methods: Possible molecular communication was assessed using in vitro assays and functional genomic approaches. Results: Our results highlight that melatonin elicits different functional microbial impacts, both at transcriptional and phenotypic levels (i.e., adhesion to intestinal cells), that are dependent on the bifidobacterial species analyzed. Conclusion: Among the bifidobacterial taxa assayed, Bifidobacterium bifidum demonstrated the highest level of molecular interaction with melatonin, highlighting its significant role in this process.
Keywords: CST; Melatonin; bifidobacteria; gut microbes; hormones.
© The Author(s) 2025.
Conflict of interest statement
Longhi G serves as Junior Editorial Board member, van Sinderen D as Co-Editor-in-Chief, Ventura M as Editor-in-Chief, and Turroni F as Executive Editor of Microbiome Research Reports. None of these individuals was involved in any aspect of the editorial process for this manuscript, including reviewer selection, manuscript handling, or decision making. The other authors declared that there are no conflicts of interest.
Figures

References
LinkOut - more resources
Full Text Sources
