MoMkt1, a member of XPG/RAD2 nuclease family, regulates development and pathogenicity in Magnaporthe oryzae
- PMID: 40852952
- PMCID: PMC12380227
- DOI: 10.1080/21505594.2025.2546068
MoMkt1, a member of XPG/RAD2 nuclease family, regulates development and pathogenicity in Magnaporthe oryzae
Abstract
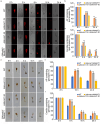
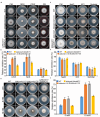
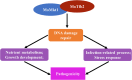
XPG/RAD2 nuclease family plays a crucial role in DNA damage repair to maintain genomic integrity. However, the biological function of Mkt1, a member of the XPG/RAD2 nuclease family, remains unclear in Magnaporthe oryzae. In this study, we identified and characterized the biological functions of MoMkt1. Our results demonstrated that MoMkt1 is involved in hyphal growth, conidiation, normal appressorium development, degradation of glycogen and lipid droplets, cell wall stress and oxidative stress responses, and pathogenicity in M. oryzae. Additionally, MoMkt1 is required for scavenging host-derived reactive oxygen species (ROS) and responding to DNA replicative stress. Further investigation revealed that MoMkt1 interacts with the transcription factor IIH (TFIIH) core subunit MoTfb2. Overexpression (OE) of MoTFB2 slightly affected the virulence of M. oryzae. Transcriptome data revealed that MoMKT1 regulates metabolic pathways, cell wall, oxidation-reduction processes, and DNA repair. Our study provides insights into MoMkt1-mediated development and pathogenicity in M. oryzae.
Keywords: DNA repair; Magnaporthe oryzae; MoMkt1; XPG/RAD2 nuclease family; pathogenicity.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures








Similar articles
-
Dihydroorotase MoPyr4 is required for development, pathogenicity, and autophagy in rice blast fungus.Cell Commun Signal. 2024 Jul 15;22(1):362. doi: 10.1186/s12964-024-01741-4. Cell Commun Signal. 2024. PMID: 39010102 Free PMC article.
-
Decarboxylase mediated oxalic acid metabolism is important to antioxidation and detoxification rather than pathogenicity in Magnaporthe oryzae.Virulence. 2025 Dec;16(1):2444690. doi: 10.1080/21505594.2024.2444690. Epub 2025 Jan 26. Virulence. 2025. PMID: 39814555 Free PMC article.
-
Decapeptide Inducer Promotes the Conidiation of Phytopathogenic Magnaporthe oryzae via the Mps1 MAPK Signaling Pathway.Int J Mol Sci. 2025 Jun 19;26(12):5880. doi: 10.3390/ijms26125880. Int J Mol Sci. 2025. PMID: 40565344 Free PMC article.
-
Golgin Protein MoCoy1 Mediates Retrograde Trafficking From the Golgi to the ER, Regulating Fungal Development and Pathogenicity in Magnaporthe oryzae.Mol Plant Pathol. 2025 Jul;26(7):e70130. doi: 10.1111/mpp.70130. Mol Plant Pathol. 2025. PMID: 40700342 Free PMC article.
-
Characterisation of guided entry of tail-anchored proteins in Magnaporthe oryzae.PLoS Pathog. 2025 Jul 28;21(7):e1013011. doi: 10.1371/journal.ppat.1013011. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40720549 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
