Gut microbiome composition and diversity of wild-caught and hatchery-bred milkfish (Chanos chanos) fry
- PMID: 40854639
- PMCID: PMC12197973
- DOI: 10.1016/j.jgeb.2025.100520
Gut microbiome composition and diversity of wild-caught and hatchery-bred milkfish (Chanos chanos) fry
Abstract
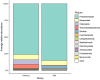
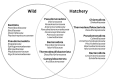
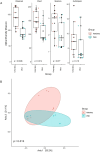
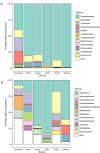
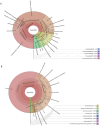
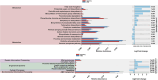
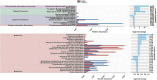
Milkfish is the most produced finfish in the Philippines, with approximately 75 % of its fry sourced from hatcheries. Despite numerous studies on gut microbiota of wild and cultured fish species, the diversity and functional roles of the milkfish fry gut microbiome remain poorly understood. This study presents the first gut microbiome profiles of wild and hatchery-bred milkfish fry using 16S rRNA amplicon analysis. A total of 437 OTUs were recovered and significant differences in gut bacterial communities among fry from different sources was observed, indicating that habitat is a key determinant of gut microbiome diversity. The core gut microbiota analysis identified Vibrionaceae and Roseobacteraceae as the most common and abundant bacterial families across fry sources. However, Paenibacillaceae and Bacillaceae under Phylum Bacillota were dominant in wild fry sources, particularly Hamtic and Kirayan, whereas families belonging to Phyla Cyanobacteriota, and Thermodesulfobacteria were more prevalent in Dumagas and Kirayan hatchery fry sources. Functional predictions of the gut bacterial microbiome revealed 26 differentially abundant pathways between wild-caught and hatchery-bred fry, including those related to metabolism, organismal systems, cellular processes, environmental and genetic information processing. These findings highlight significant variations in gut microbiome composition, diversity, and functional potential across different sources of wild-caught and hatchery-bred fry. Understanding these source-specific microbial communities could provide insight into the development of interventions that can improve gut health and enhance milkfish hatchery practices. It can also generate information on ideal fry selection across local milkfish sources that will enhance larval productivity and survival in the succeeding nursery and grow-out culture stages.
Keywords: Gut microbiome; Hatchery-bred; Metagenome; Microbial diversity; Milkfish fry; Wild-caught.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Philippine Statistics Authority. Fisheries Situationer Report: July to September 2024. Published online November 2024. https://psa.gov.ph/system/files/fsd/Fisheries%20Situation%20Report%2C%20....
-
- 2023 Philippine Fisheries Profile. Published online October 2024. https://www.bfar.da.gov.ph/wp-content/uploads/2025/02/2023-Philippine-Fi....
-
- Hilomen-Garcia G.V. Morphological abnormalities in hatchery-bred milkfish (Chanos chanos forsskal) fry and juveniles. Aquaculture. 1997;152(1–4):155–166. doi: 10.1016/S0044-8486(96)01518-9. - DOI
-
- Bureau of Fisheries and Aquatic Resources. National Milkfish Industry Roadmap 2021-2040.; 2022. https://www.bfar.da.gov.ph/wp-content/uploads/2022/11/Milkfish-Industry-....
-
- Baliao D, Navarro J, Tugo JA, Santiagudo EM. Towards Increased Production of Milkfish Fry in the Philippines: SEAFDEC/AQD technology through the lens. 2021;19(2).
LinkOut - more resources
Full Text Sources
