DNA double-strand break end resection factors and WRN facilitate mitotic DNA synthesis in human cells
- PMID: 40854910
- PMCID: PMC12379236
- DOI: 10.1038/s41467-025-63292-7
DNA double-strand break end resection factors and WRN facilitate mitotic DNA synthesis in human cells
Abstract
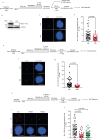
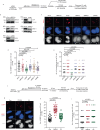
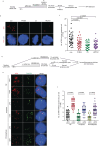
Mitotic DNA synthesis (MiDAS) serves to complete the replication of genomic loci that are not fully replicated in S phase in response to replication stress. Previous studies suggest that MiDAS might proceed via break-induced DNA replication, a sub-pathway of homologous recombination repair activated at broken or collapsed replication forks. We set out to define whether DNA double strand break end-resection factors play a role in MiDAS. Here, we show that several core end-resection factors, including MRE11, CtIP and BRCA1 are essential for MiDAS. In addition, while loss of WRN or DNA2 impairs MiDAS, there is no requirement for other known end-resection factors such as EXO1 and BLM. Moreover, both the exonuclease and the helicase activities of WRN contribute to MiDAS. Because oncogene-induced replication stress is common in cancers, targeting of WRN or other factors required for MiDAS could facilitate the development of targeted cancer therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
Promotion of DNA end resection by BRCA1-BARD1 in homologous recombination.Nature. 2024 Oct;634(8033):482-491. doi: 10.1038/s41586-024-07910-2. Epub 2024 Sep 11. Nature. 2024. PMID: 39261729 Free PMC article.
-
DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells.J Biol Chem. 2014 Sep 26;289(39):27314-27326. doi: 10.1074/jbc.M114.578823. Epub 2014 Aug 13. J Biol Chem. 2014. PMID: 25122754 Free PMC article.
-
DNA2 drives processing and restart of reversed replication forks in human cells.J Cell Biol. 2015 Mar 2;208(5):545-62. doi: 10.1083/jcb.201406100. J Cell Biol. 2015. PMID: 25733713 Free PMC article.
-
Mechanism of BRCA1-BARD1 function in DNA end resection and DNA protection.Nature. 2024 Oct;634(8033):492-500. doi: 10.1038/s41586-024-07909-9. Epub 2024 Sep 11. Nature. 2024. PMID: 39261728 Free PMC article.
-
Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template.Biochem Cell Biol. 2007 Aug;85(4):509-20. doi: 10.1139/O07-069. Biochem Cell Biol. 2007. PMID: 17713585 Review.
References
MeSH terms
Substances
Grants and funding
- DNRF 115/Danmarks Grundforskningsfond (Danish National Research Foundation)
- Antihelix 859853/EC | EC Seventh Framework Programm | FP7 People: Marie-Curie Actions (FP7-PEOPLE - Specific Programme "People" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- 1030-00180B/Natur og Univers, Det Frie Forskningsråd (Natural Sciences, Danish Council for Independent Research)
LinkOut - more resources
Full Text Sources
Miscellaneous

