ERH promotes primary microRNA processing beyond cluster assistance
- PMID: 40854975
- PMCID: PMC12379101
- DOI: 10.1038/s41467-025-63015-y
ERH promotes primary microRNA processing beyond cluster assistance
Abstract
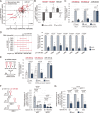
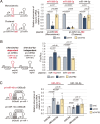
MicroRNA (miRNA) maturation is initiated by the Microprocessor complex, comprising DROSHA and DGCR8, that processes primary miRNAs (pri-miRNAs). Recent studies have identified ERH and SAFB2 as auxiliary factors that enhance the functionality of the Microprocessor. These factors are required for cluster assistance, where optimal pri-miRNAs facilitate the processing of adjacent suboptimal pri-miRNAs. However, the specific action mechanisms of ERH and SAFB2 have not yet been defined. In this study, we found that ERH broadly enhances the processing of pri-miRNAs regardless of their genomic contexts, affecting both stand-alone and clustered ones. Suboptimal hairpins are affected more prominently by ERH knockdown than efficiently processed hairpins. In contrast, SAFB2 specifically supports the processing of suboptimal pri-miRNA hairpins within clusters. This study reveals the distinct roles of ERH and SAFB2 in cluster assistance and presents a new model, in which SAFB2 facilitates the Microprocessor's transfer between hairpins, while ERH enables the efficient processing of suboptimal pri-miRNAs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

