The HDAC inhibitor romidepsin renders liver cancer vulnerable to RTK targeting and immunologically active
- PMID: 40855049
- PMCID: PMC12378214
- DOI: 10.1038/s41467-025-62934-0
The HDAC inhibitor romidepsin renders liver cancer vulnerable to RTK targeting and immunologically active
Abstract
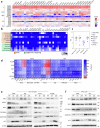
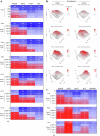
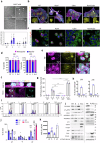
Histone deacetylases (HDACs) are epigenetic regulators frequently altered in cancer. Here we report that overexpression of HDAC1/2 occurs in Hepatocellular Carcinoma (HCC) patients, correlating with poor prognosis. We show that romidepsin, a class-I HDAC inhibitor, elicits a combinatorial perturbation of distinct molecular processes in HCC cells, altering lipid composition, mitotic spindle machinery, and levels of cell cycle/survival signals. Collectively, these alterations lead HCC cells to a vulnerable state, conferring dependency to receptor tyrosine kinase (RTK) signalling support. The cytostatic effects of romidepsin alone is converted into cytotoxicity by the RTK inhibitor cabozantinib in HCC models. We document that romidepsin+cabozantibib confers an immune-stimulatory profile in Alb-R26Met mouse models, with direct effects on primary human dendritic cell maturation in vitro. Our findings put forward the intricate crosstalk between epigenetics, metabolism, and immune response in cancer. The broad action of romidepsin on distinct cellular functions highlights its therapeutic potential for HCC treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Yang, F. F., Hu, T., Liu, J. Q., Yu, X. Q. & Ma, L. Y. Histone deacetylases (HDACs) as the promising immunotherapeutic targets for hematologic cancer treatment. Eur. J. Med Chem.245, 114920 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

