Fishing out AIEC with FimH capturing microgels for inflammatory bowel disease treatment
- PMID: 40855058
- PMCID: PMC12378211
- DOI: 10.1038/s41467-025-63276-7
Fishing out AIEC with FimH capturing microgels for inflammatory bowel disease treatment
Abstract
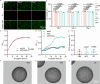
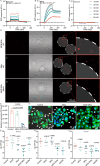
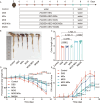
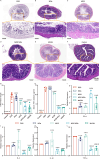
Inflammatory bowel disease (IBD) is a chronic immune-mediated condition with rising global incidence and limited treatment options. Current therapies often have poor efficacy and undesirable side effects. Here we present a drug-free strategy that targets bacterial adhesion to manage IBD. We develop porous microgels loaded with mannan oligosaccharides (MOS) that mimic the natural binding sites of intestinal cells. These microgels attract adherent-invasive Escherichia coli (AIEC) by interacting with FimH, a bacterial protein used for attachment, thereby preventing AIEC from colonizing the gut lining. The microgels are fabricated using an all-aqueous two-phase system, enabling biocompatibility and structural control. In a mouse model of IBD, this competitive adsorption approach alleviates intestinal inflammation, reduces harmful Enterobacteriaceae, and enhances gut microbial diversity. This work introduces a non-antibiotic, bioinspired method that intercepts pathogenic bacteria and restores microbial balance, offering a promising therapeutic strategy for IBD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors have thoroughly reviewed the journal’s policy on Competing Interests and declare that they have no competing interests related to this research.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

