Modeling of T cell-mediated autoimmune pituitary disease using human induced pluripotent stem cell-originated organoid
- PMID: 40855071
- PMCID: PMC12378982
- DOI: 10.1038/s41467-025-63183-x
Modeling of T cell-mediated autoimmune pituitary disease using human induced pluripotent stem cell-originated organoid
Abstract
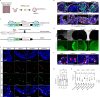
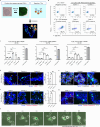
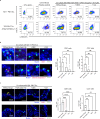
Anti-pituitary-specific transcription factor (PIT)-1 hypophysitis is an autoimmune disease characterized by hormone secretion impairment from PIT-1-expressing pituitary cells, accompanied by malignancies with ectopic PIT-1 expression. Cytotoxic T cells (CTL) targeting PIT-1-positive cells have been implicated in disease development, yet direct evidence is lacking. As human leukocyte antigen (HLA)-matching is required for modeling T cell-mediated autoimmune diseases, we employ induced pluripotent stem cells (iPSC) to generate pituitary organoids harboring the patients' HLA haplotype and coculture the organoids with PIT-1-reactive CTLs isolated from the patients' peripheral blood mononuclear cells. The coculture demonstrates specific CTL-mediated cytotoxicity against PIT-1-positive cells exclusively in autologous conditions, with this cytotoxicity inhibited by immunosuppressive agents such as dexamethasone and cyclosporin A. Multiple combinations of epitopes, CTLs, and HLA molecules are responsible for pathogenesis. These data demonstrate CTL-mediated autoimmunity in anti-PIT-1 hypophysitis and highlight the potential application of this strategy for other T cell-mediated autoimmune diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Yutaka Takahashi has received research funding from Teijin Pharma Co., Ltd. and Ono Pharma Co., Ltd. Shin Kaneko is a founder, shareholder, and director of Thyas Co., Ltd. and has received research funding from Takeda Pharmaceutical Co., Ltd., Astellas Co., Ltd., Kirin Co., Ltd., Terumo Co., Ltd., and Thyas Co., Ltd. The other authors have nothing to declare.
Figures


References
-
- Yamamoto, M. et al. Autoimmune pituitary disease: new concepts with clinical implications. Endocr. Rev.41, bnz003 (2020). - PubMed
-
- Takahashi, Y. Mechanisms in endocrinology: autoimmune hypopituitarism: novel mechanistic insights. Eur. J. Endocrinol.182, R59–r66 (2020). - PubMed
-
- Bando, H. et al. Paraneoplastic autoimmune hypophysitis: an emerging concept. Best. Pract. Res. Clin. Endocrinol. Metab.36, 101601 (2021). - PubMed
-
- Bando, H. et al. Involvement of PIT-1-reactive cytotoxic T lymphocytes in anti-PIT-1 antibody syndrome. J. Clin. Endocrinol. Metab.99, E1744–E1749 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

