High throughput tear proteomics with data independent acquisition enables biomarker discovery in allergic conditions
- PMID: 40855359
- PMCID: PMC12379103
- DOI: 10.1038/s41598-025-17105-y
High throughput tear proteomics with data independent acquisition enables biomarker discovery in allergic conditions
Abstract
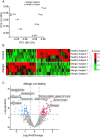
The search for pathological biomarkers in biological fluids that can provide valuable insight into an individual's health status, is a relevant area of research for multiple pathologies. Currently, the use of proteomics for the identification of differences in protein expression profiles between samples from healthy subjects and patients, has emerged as a powerful strategy to improve the current diagnosis of various pathologies or propose novel therapeutic approaches. Among the biological fluids from which new pathological biomarkers can be identified, tear secretion is highly attractive, since it can be collected non-invasively and could better concentrate proteins that sensitively reflect allergic responses, owing to their exposure to environmental factors and its connection to the respiratory system. Despite its potential, tear fluid remains underexplored, offering significant research opportunities. In this study, we collected human tear samples using the Shirmer Test from healthy and allergic individuals. Our optimized workflow, combining sample preparation and high-throughput proteomics using the data-independent acquisition (DIA) strategy, identified 2542 proteins and enabled the successful differentiation of the two groups. We identified 99 differentially expressed proteins. Our results show the feasibility of protein analysis in human tear samples, highlighting tears as a highly sensitive fluid for detecting health conditions. Data are available via ProteomeXchange with identifier PXD067099.
Keywords: Biomarkers; DiaPASEF; High-throughput proteomics; Tear secretion.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Word Health Organization. Strengthening diagnostics capacity. Resolution from executive board 152nd session, Agenda item 5. (2023).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

