Senescent fibroblasts secrete CTHRC1 to promote cancer stemness in hepatocellular carcinoma
- PMID: 40855439
- PMCID: PMC12376455
- DOI: 10.1186/s12964-025-02369-8
Senescent fibroblasts secrete CTHRC1 to promote cancer stemness in hepatocellular carcinoma
Abstract
Background: Cellular senescence plays a significant role in tumorigenesis and tumor progression. Substantial evidence indicates that senescence occurs in cancer-associated fibroblasts (CAFs), the predominant stromal component within the tumor microenvironment (TME), which profoundly impacts tumor biology. However, despite growing evidence of stromal cell involvement in cancer progression, the specific mechanisms and clinical implications of senescent CAFs (SCAFs) in hepatocellular carcinoma (HCC) have not been fully elucidated.
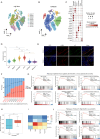
Methods: The senescence signature was utilized to evaluate the senescence status of cell types within the TME of HCC using the GSE149614 dataset. The CytoTRACE and cell-cell communication analysis were used to find the correlation between cancer stemness and SCAFs. A risk prediction model associated with SCAFs was constructed to investigate potential mechanisms by which SCAFs promote tumor progression. Single-cell RNA sequencing data was used to identify senescent CAF-related genes. Gene expression and clinical data for HCC were obtained from the Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), and National Omics Data Encyclopedia (NODE) databases. Using four machine-learning algorithms, crucial genes were identified to develop a CAF-senescence-related risk model, predicting prognosis, cancer stemness, immune infiltration, tumor mutation burden, and therapeutic responses in HCC patients. Next, we explored the role of Collagen Triple Helix Repeat Containing-1 (CTHRC1) in cancer stemness using both in vitro and in vivo experiments. Through various functional experiments, we elucidated the downstream signaling pathways of CTHRC1. Additionally, chromatin immunoprecipitation experiments were used to verify that key transcription factors bind to the CTHRC1 promoter region.
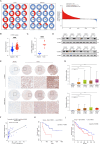
Results: CAFs exhibited high senescence status and a strong correlation with cancer stemness in HCC. A novel CAF-senescence-score (CSscore) prognostic model was established for HCC based on 10 genes: CTHRC1, SERPINE1, RNF11, ENG, MARCKSL1, ASAP1, FHL3, LAMB1, CD151, and OLFML2B. The survival prediction performance was validated on TCGA, ICGC, and NODE cohorts. Immune analysis revealed that the CSscore was positively correlated with immunosuppressive immune cell populations, including M2 macrophages and regulatory T cells. Conversely, a negative correlation was observed between the CSscore and anti-tumor immune cells such as CD8 + T cells, dendritic cells, and B cells HCC patients with a low CSscore had a lower tumor mutation burden and showed improved responsiveness to immunotherapy and transarterial chemoembolization. In vitro experiments and bioinformatics analysis further revealed that CTHRC1 was significantly elevated in SCAFs promoted cancer stemness and metastasis via the SRY-box transcription factor 4 (SOX4)-CTHRC1-Notch1 axis in HCC.
Conclusion: Our study revealed that SCAFs were strongly correlated with cancer stemness in HCC. A novel machine learning model based on senescent CAF-related genes was constructed to accurately predict prognosis in HCC patients. Furthermore, CTHRC1 was identified as a novel prognostic and therapeutic biomarker to predict poor prognosis in HCC and promote cancer stemness and metastasis through the Notch signaling pathway, with its expression being transcriptionally regulated by SOX4.
Keywords: Cancer stemness; Cancer-associated fibroblasts; Cellular senescence; Hepatocellular carcinoma; Notch signaling pathway.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The TCGA and GEO databases are publicly available. Users can download the data for free for research purposes and publish related articles. The present study for animal experiments was approved by the Institutional Animal Care and Use Committee, Huazhong University of Science and Technology (HUST), Wuhan, China. The present study for collecting clinical samples was approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology (HUST), Wuhan, China. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures








References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14. - PubMed
-
- Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, Wang C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20:203–22. - PubMed
-
- Lee TK, Guan XY, Ma S. Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications. Nat Rev Gastroenterol Hepatol. 2022;19:26–44. - PubMed
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

