SLFN11, far from being limited to responding to cancer DNA damage
- PMID: 40856812
- PMCID: PMC12380651
- DOI: 10.1007/s10238-025-01776-y
SLFN11, far from being limited to responding to cancer DNA damage
Abstract
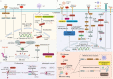
SLFN11, a member of the evolutionarily conserved SLFN gene family, is an interferon-stimulated early response gene. This review comprehensively explores its multifaceted roles. Structurally, its three distinct domains endow it with diverse functions. Epigenetic modifications, post-translational alterations, and multiple signaling pathways intricately regulate SLFN11 expression and activity. In terms of functions, it plays crucial roles in the DNA damage response during replication stress, distinct from traditional pathways. It also serves as a protector in the antiviral response and a valuable biomarker for predicting the efficacy of DNA-damaging agents and patient prognosis in various cancers. Beyond these, SLFN11 has non-canonical functions, including immune regulation, modulation of oncological behaviors, involvement in apoptosis, protection against proteotoxic stress, and association with Fanconi anemia. Looking ahead, SLFN11 holds great promise as a biomarker for personalized medicine, but challenges like developing accurate detection methods remain. In immunotherapy, understanding its dynamic changes is essential for optimizing treatment. Strategies to overcome SLFN11-low expression, such as epigenetic modulation, also need further investigation, which may open new avenues for disease treatment.
Keywords: Antiviral function; Biomarker; DNA damage response; Epigenetic modification; Immune regulation; Oncological behavior; Schlafen 11.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interests: The authors declare no competing interests. Ethical approval: Not applicable. Consent to participate: Not applicable. Consent for publication: Not applicable.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
From predictive biomarker to therapeutic target: the dual role of SLFN11 in chemotherapy sensitivity.Cancer Chemother Pharmacol. 2025 Jun 18;95(1):60. doi: 10.1007/s00280-025-04781-w. Cancer Chemother Pharmacol. 2025. PMID: 40531330 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
SLFN11: a pan-cancer biomarker for DNA-targeted drugs sensitivity and therapeutic strategy guidance.Front Oncol. 2025 Jul 22;15:1582738. doi: 10.3389/fonc.2025.1582738. eCollection 2025. Front Oncol. 2025. PMID: 40766331 Free PMC article. Review.
-
The role of SLFN11 in DNA replication stress response and its implications for the Fanconi anemia pathway.DNA Repair (Amst). 2024 Sep;141:103733. doi: 10.1016/j.dnarep.2024.103733. Epub 2024 Jul 24. DNA Repair (Amst). 2024. PMID: 39096698 Review.
References
-
- Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9(5):657–68. - PubMed
-
- Brady G, Boggan L, Bowie A, O’Neill LA. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem. 2005;280(35):30723–34. - PubMed
-
- Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370(1):62–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

