Single-cell spatial transcriptomics unravels the cellular landscape of abdominal aortic aneurysm
- PMID: 40857411
- PMCID: PMC12406718
- DOI: 10.1172/jci.insight.190534
Single-cell spatial transcriptomics unravels the cellular landscape of abdominal aortic aneurysm
Abstract
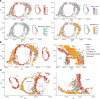
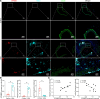
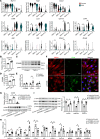
Abdominal aortic aneurysm (AAA) is a life-threatening vascular disease with no effective pharmacological interventions. While single-cell transcriptomics has advanced our understanding of AAA, it lacks spatial context. Here, we employed Seq-Scope, an ultra-high-resolution spatial transcriptomic technology, to decipher the spatial landscape of angiotensin II-induced AAA in Apoe-/- mice. Our analysis revealed the heterogeneity of macrophages, fibroblasts, and smooth muscle cells (SMCs), with specific responses in different layers of the AAA tissue. SMCs in the inner layers showed associations with Mgp-expressing fibroblasts and GPNMB-expressing macrophages, whereas the outer layers had different dominant cell types. Notably, GPNMB-expressing macrophages were concentrated near SMCs in regions of severe elastic lamina damage. Immunofluorescent staining confirmed their colocalization, and scRNA-seq reanalysis independently validated the presence of GPNMB-high macrophages in AAA tissues, highlighting their involvement in inflammation and tissue remodeling. Moreover, we discovered that macrophage-derived soluble GPNMB induces SMC phenotypic switching, reducing contractile markers while increasing cytokines and metalloproteinases. This effect was partly mediated by CD44 signaling. These findings suggest that GPNMB-high macrophages contribute to AAA development by driving SMC dysfunction. This study highlights the importance of high-resolution spatial transcriptomics in complementing single-cell transcriptomics, offering valuable insights into molecular and cellular responses in the AAA microenvironment.
Keywords: Cardiology; Cardiovascular disease; Vascular biology.
Figures

References
MeSH terms
Substances
Grants and funding
- R01 HL162294/HL/NHLBI NIH HHS/United States
- R01 AG079163/AG/NIA NIH HHS/United States
- R01 HL151776/HL/NHLBI NIH HHS/United States
- UG3 CA268091/CA/NCI NIH HHS/United States
- R01 HL134569/HL/NHLBI NIH HHS/United States
- U01 HL137182/HL/NHLBI NIH HHS/United States
- R01 HL172832/HL/NHLBI NIH HHS/United States
- R01 HL138139/HL/NHLBI NIH HHS/United States
- UH3 CA268091/CA/NCI NIH HHS/United States
- R01 HL109946/HL/NHLBI NIH HHS/United States
- R01 HL153710/HL/NHLBI NIH HHS/United States
- R01 HL141891/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

