Tumor-derived PRMT1 suppresses macrophage antitumor activity by inhibiting cGAS/STING signaling in gastric cancer cells
- PMID: 40858547
- PMCID: PMC12381180
- DOI: 10.1038/s41419-025-07960-y
Tumor-derived PRMT1 suppresses macrophage antitumor activity by inhibiting cGAS/STING signaling in gastric cancer cells
Abstract
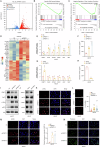
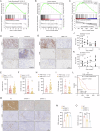
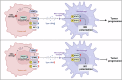
Gastric cancer (GC) is a common and aggressive malignancy worldwide. Increasing evidence has shown that epigenetic changes are closely related to the development of cancer and tumor-associated macrophages. Here, we report that PRMT1 is a key immunosuppressive factor in GC. PRMT1 is upregulated in GC and promotes tumor progression. PRMT1 knockdown in GC leads to the activation of the cGAS/STING pathway through the enhancement of dsDNA aggregation, which subsequently increases IFN-β secretion. Notably, after PRMT1 knockdown, M1-like tumor-associated macrophage (TAM) infiltration increased, whereas M2-like TAM infiltration decreased in vivo and in vitro. After the targeted inhibition of STING by siRNA or H151, the improvement in the progression of GC caused by PRMT1 knockdown decreased, and the changes in macrophage polarization were reversed. Furthermore, we found that PRMT1 knockdown in GC affects the STAT pathway in TAMs, inducing changes in their polarization and promoting GC apoptosis by enhancing IFN-β secretion through the cGAS/STING pathway. In summary, our findings revealed that PRMT1 knockdown inhibits the cGAS/STING pathway in GC, which produces type I IFNs to promote the polarization of M1-like macrophages in the tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA: Cancer J Clin. 2021;71:7–33. - PubMed
-
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet (Lond, Engl). 2020;396:635–48. - PubMed
-
- Chin EN, Yu C, Vartabedian VF, Jia Y, Kumar M, Gamo AM, et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science (N. Y, NY). 2020;369:993–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

